healthyverse_tsa
Time Series Analysis, Modeling and Forecasting of the Healthyverse
Packages Steven P. Sanderson II, MPH - Date: 2026-02-06
Introduction
This analysis follows a Nested Modeltime Workflow from modeltime
along with using the NNS package. I use this to monitor the
downloads of all of my packages:
Get Data
glimpse(downloads_tbl)
Rows: 167,694
Columns: 11
$ date <date> 2020-11-23, 2020-11-23, 2020-11-23, 2020-11-23, 2020-11-23,…
$ time <Period> 15H 36M 55S, 11H 26M 39S, 23H 34M 44S, 18H 39M 32S, 9H 0M…
$ date_time <dttm> 2020-11-23 15:36:55, 2020-11-23 11:26:39, 2020-11-23 23:34:…
$ size <int> 4858294, 4858294, 4858301, 4858295, 361, 4863722, 4864794, 4…
$ r_version <chr> NA, "4.0.3", "3.5.3", "3.5.2", NA, NA, NA, NA, NA, NA, NA, N…
$ r_arch <chr> NA, "x86_64", "x86_64", "x86_64", NA, NA, NA, NA, NA, NA, NA…
$ r_os <chr> NA, "mingw32", "mingw32", "linux-gnu", NA, NA, NA, NA, NA, N…
$ package <chr> "healthyR.data", "healthyR.data", "healthyR.data", "healthyR…
$ version <chr> "1.0.0", "1.0.0", "1.0.0", "1.0.0", "1.0.0", "1.0.0", "1.0.0…
$ country <chr> "US", "US", "US", "GB", "US", "US", "DE", "HK", "JP", "US", …
$ ip_id <int> 2069, 2804, 78827, 27595, 90474, 90474, 42435, 74, 7655, 638…
The last day in the data set is 2026-02-04 22:18:40, the file was birthed on: 2025-10-31 10:47:59.603742, and at report knit time is 2311.51 hours old. Happy analyzing!
Now that we have our data lets take a look at it using the skimr
package.
skim(downloads_tbl)
| Name | downloads_tbl |
| Number of rows | 167694 |
| Number of columns | 11 |
| _______________________ | |
| Column type frequency: | |
| character | 6 |
| Date | 1 |
| numeric | 2 |
| POSIXct | 1 |
| Timespan | 1 |
| ________________________ | |
| Group variables | None |
Data summary
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| r_version | 123618 | 0.26 | 5 | 7 | 0 | 50 | 0 |
| r_arch | 123618 | 0.26 | 1 | 7 | 0 | 6 | 0 |
| r_os | 123618 | 0.26 | 7 | 19 | 0 | 24 | 0 |
| package | 0 | 1.00 | 7 | 13 | 0 | 8 | 0 |
| version | 0 | 1.00 | 5 | 17 | 0 | 63 | 0 |
| country | 15699 | 0.91 | 2 | 2 | 0 | 166 | 0 |
Variable type: Date
| skim_variable | n_missing | complete_rate | min | max | median | n_unique |
|---|---|---|---|---|---|---|
| date | 0 | 1 | 2020-11-23 | 2026-02-04 | 2023-12-03 | 1893 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| size | 0 | 1 | 1125724.70 | 1484445.10 | 355 | 37869 | 322853 | 2348316 | 5677952 | ▇▁▂▁▁ |
| ip_id | 0 | 1 | 11204.87 | 21833.24 | 1 | 221 | 2790 | 11717 | 299146 | ▇▁▁▁▁ |
Variable type: POSIXct
| skim_variable | n_missing | complete_rate | min | max | median | n_unique |
|---|---|---|---|---|---|---|
| date_time | 0 | 1 | 2020-11-23 09:00:41 | 2026-02-04 22:18:40 | 2023-12-03 06:45:13 | 106185 |
Variable type: Timespan
| skim_variable | n_missing | complete_rate | min | max | median | n_unique |
|---|---|---|---|---|---|---|
| time | 0 | 1 | 0 | 59 | 52 | 60 |
We can see that the following columns are missing a lot of data and for
us are most likely not useful anyways, so we will drop them
c(r_version, r_arch, r_os)
Plots
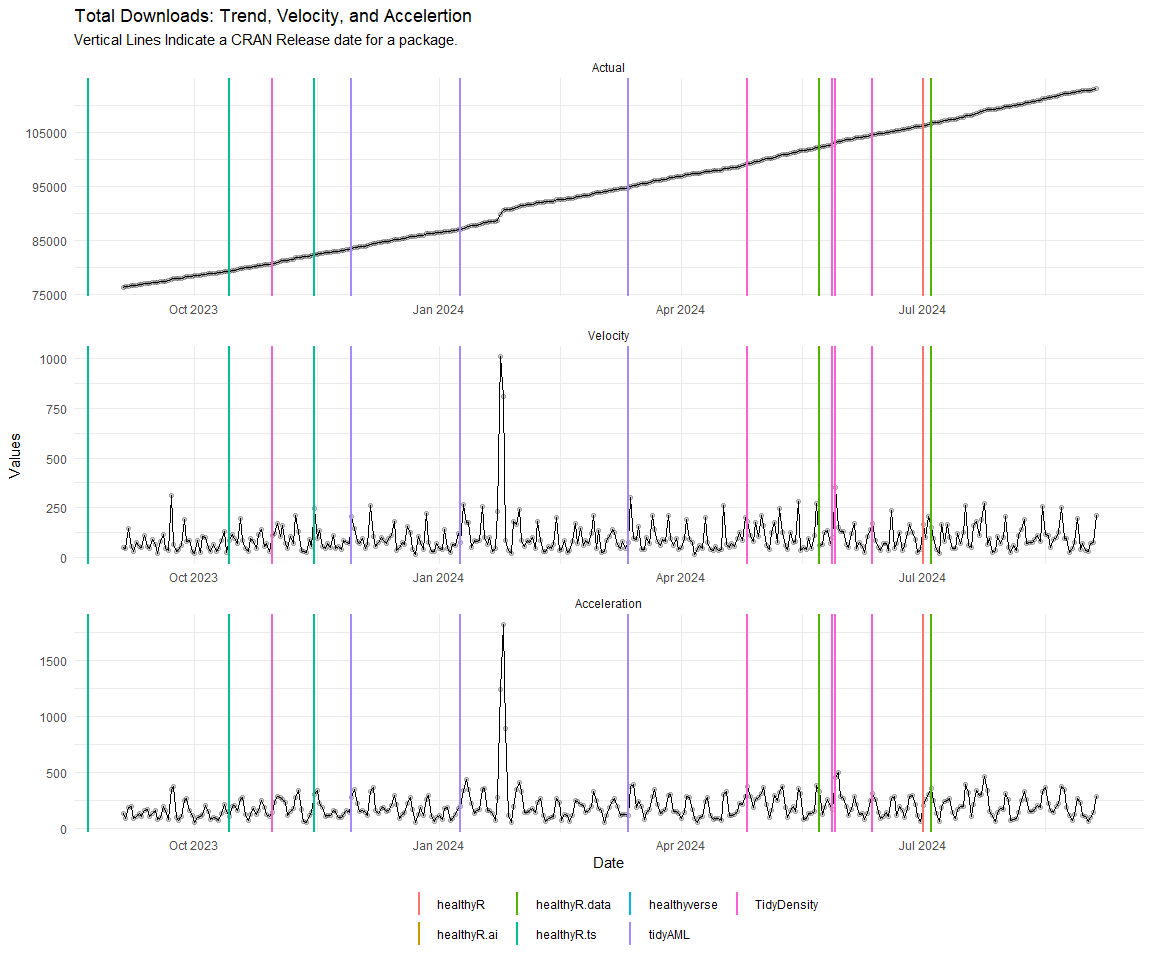
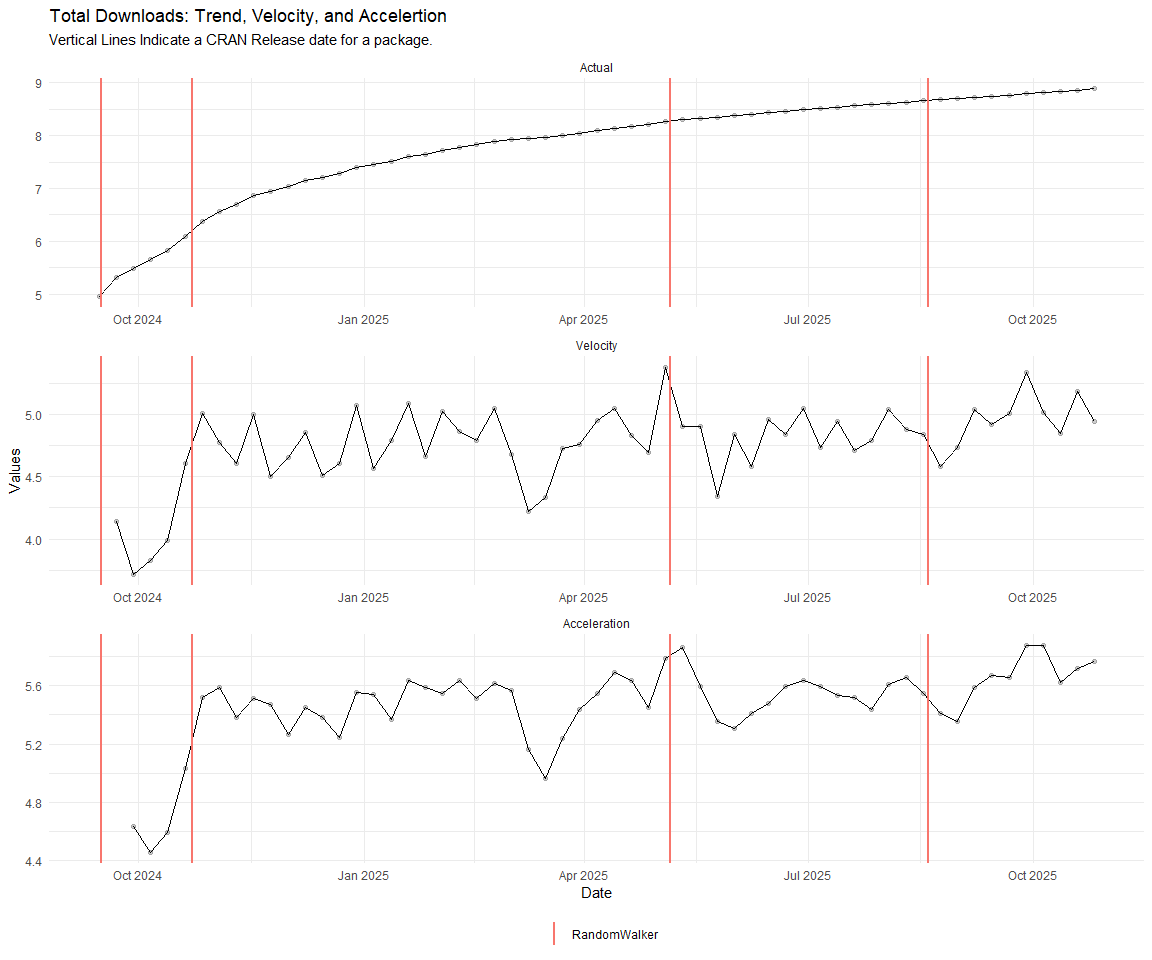
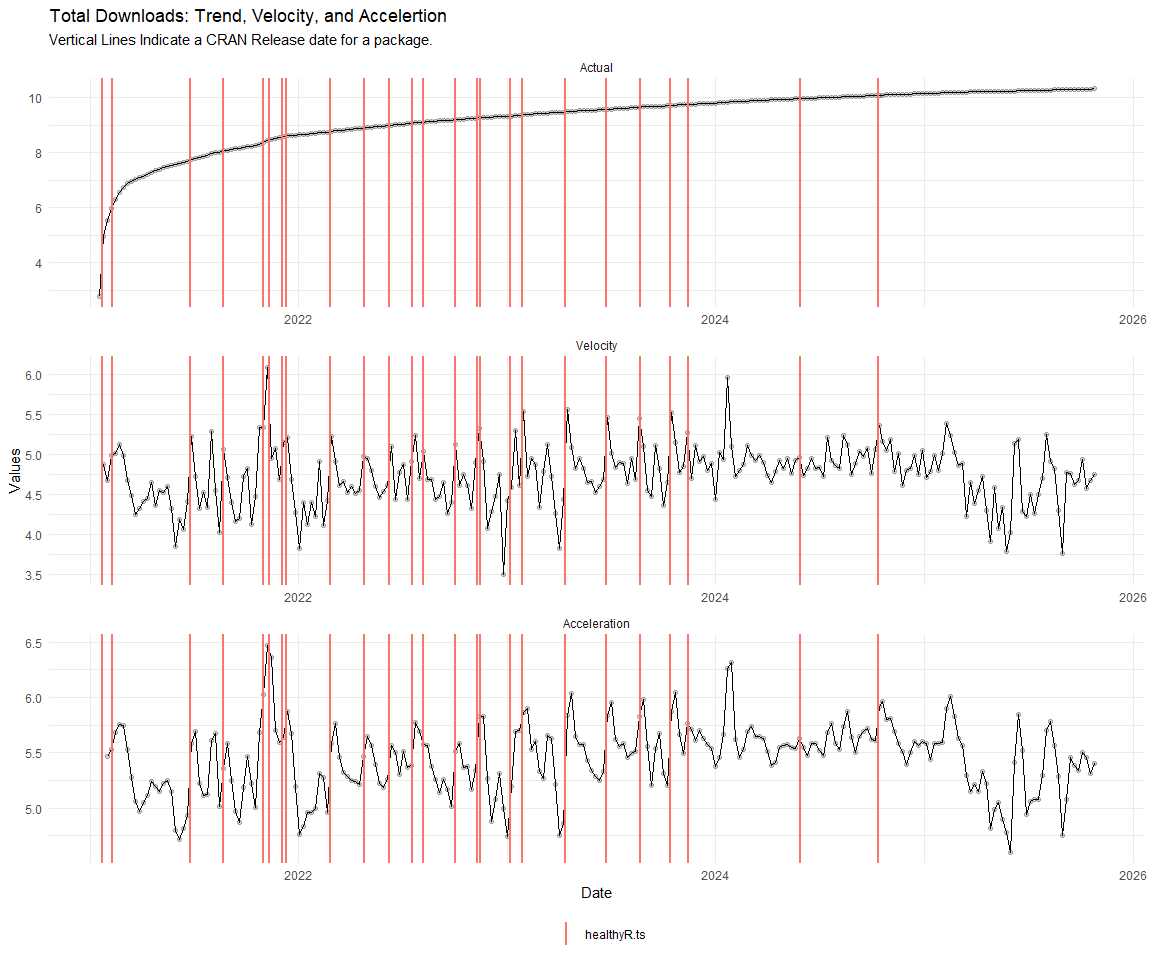
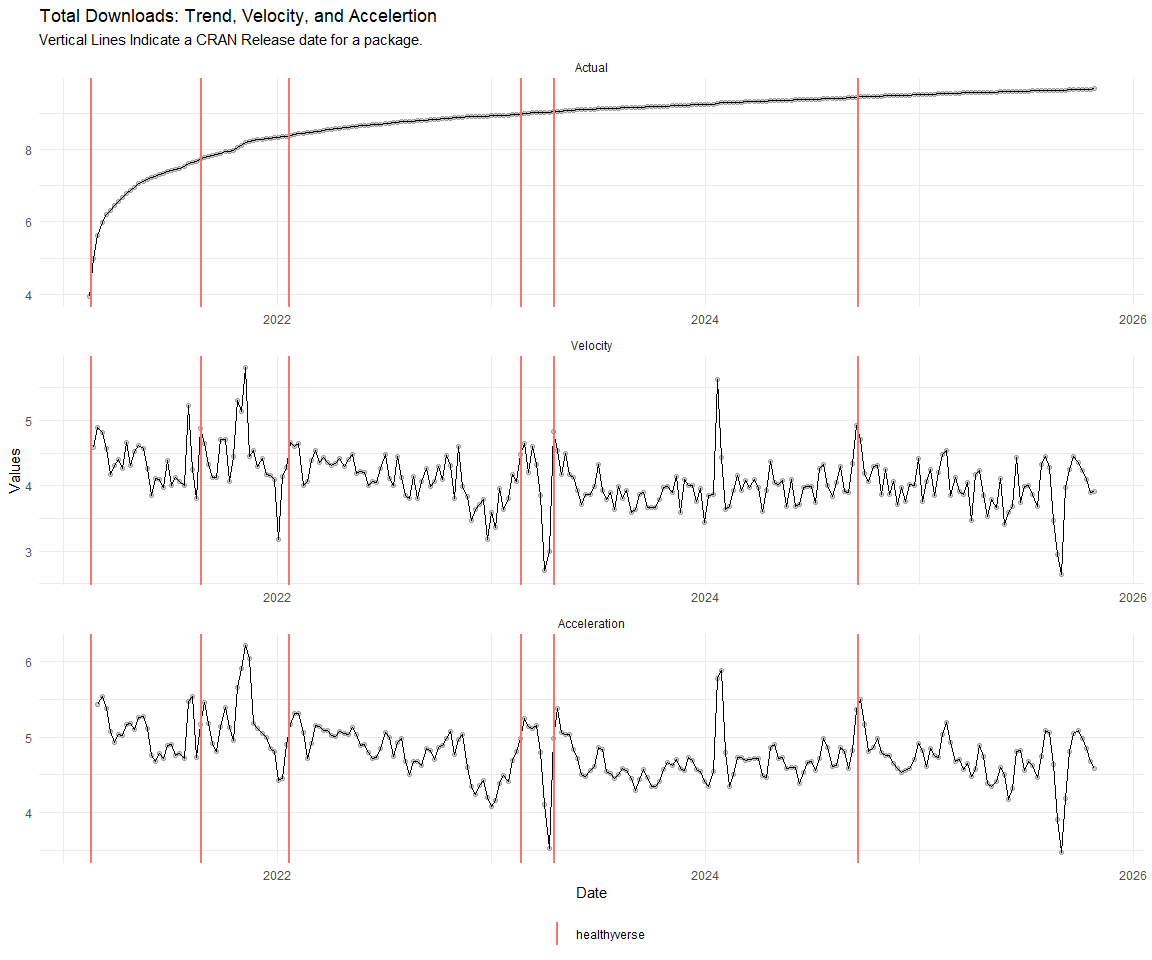
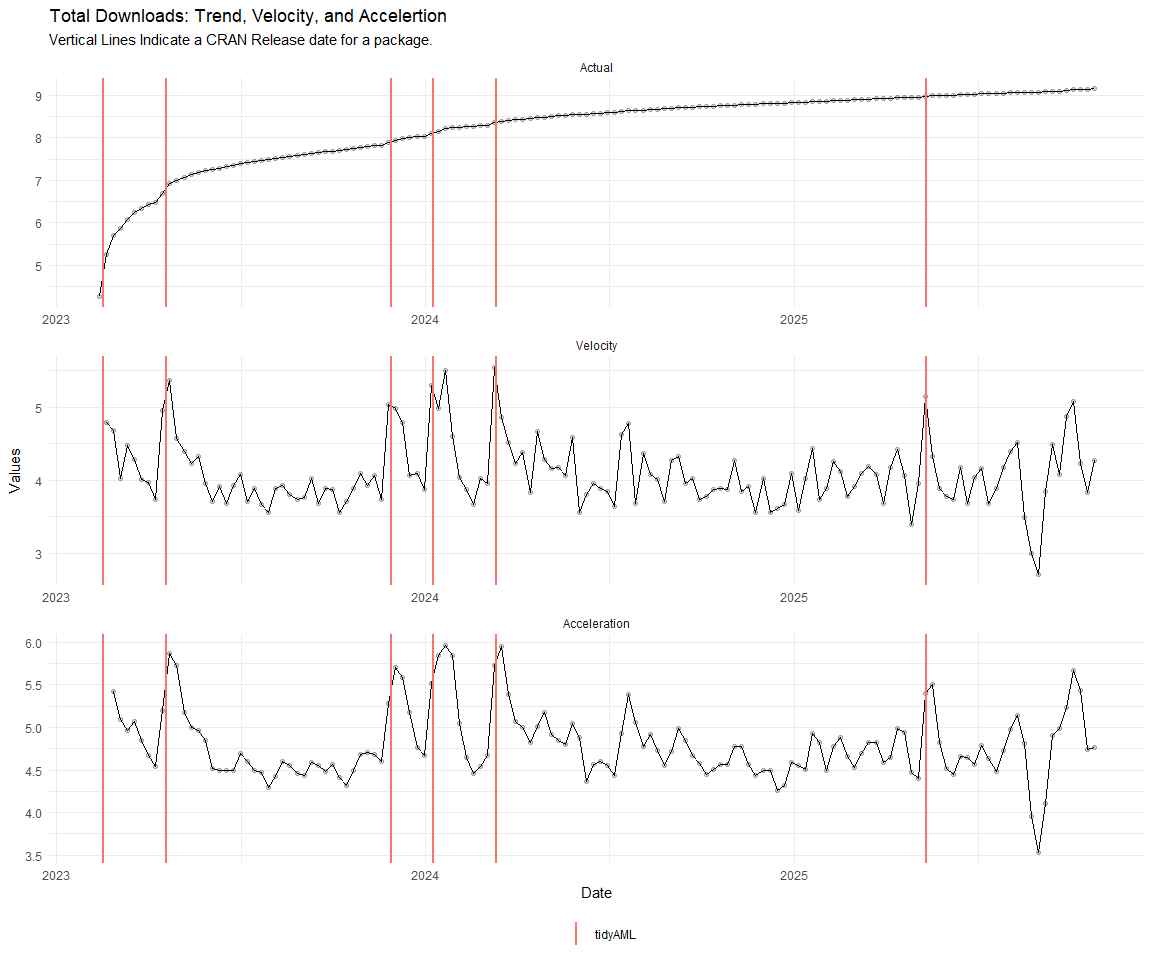
Now lets take a look at a time-series plot of the total daily downloads by package. We will use a log scale and place a vertical line at each version release for each package.
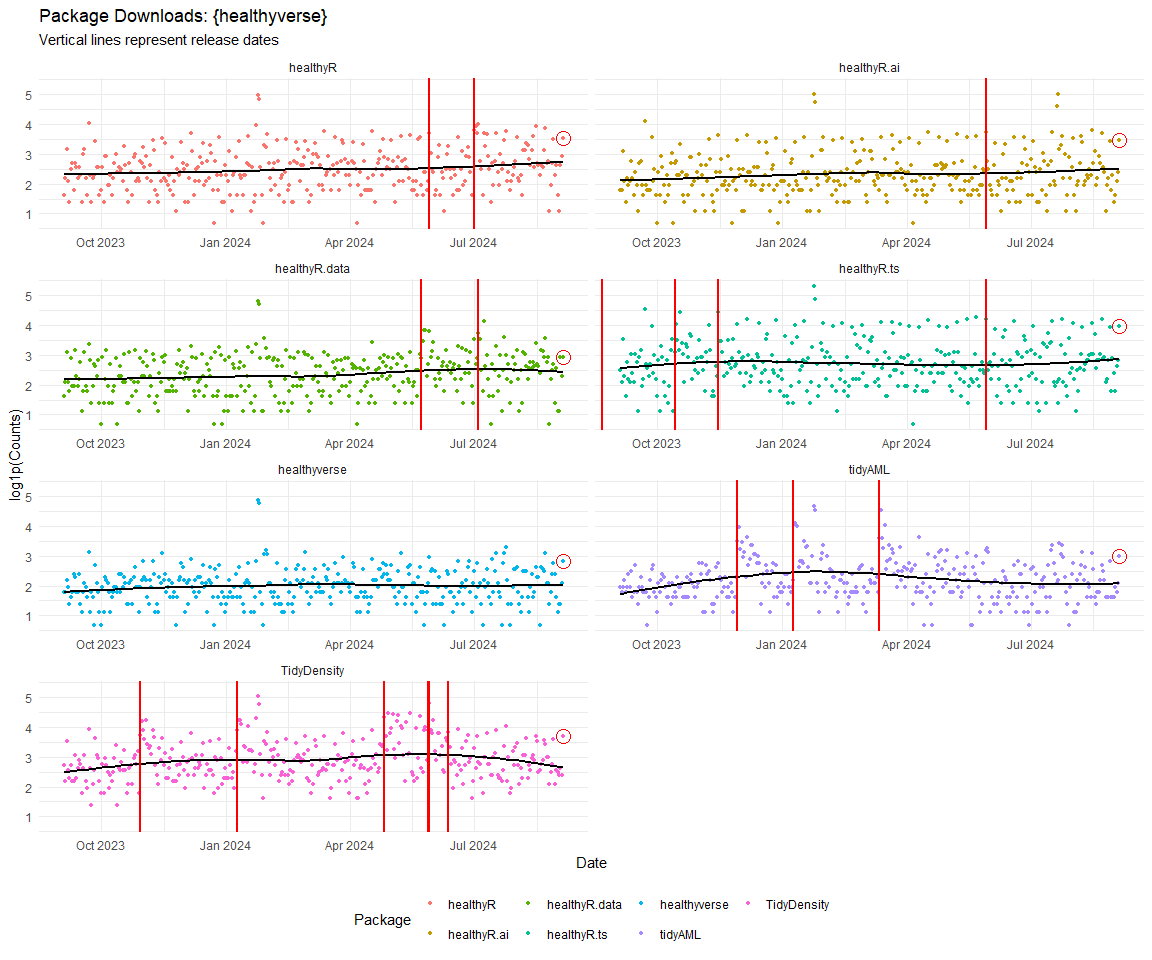
[[1]]
[[2]]
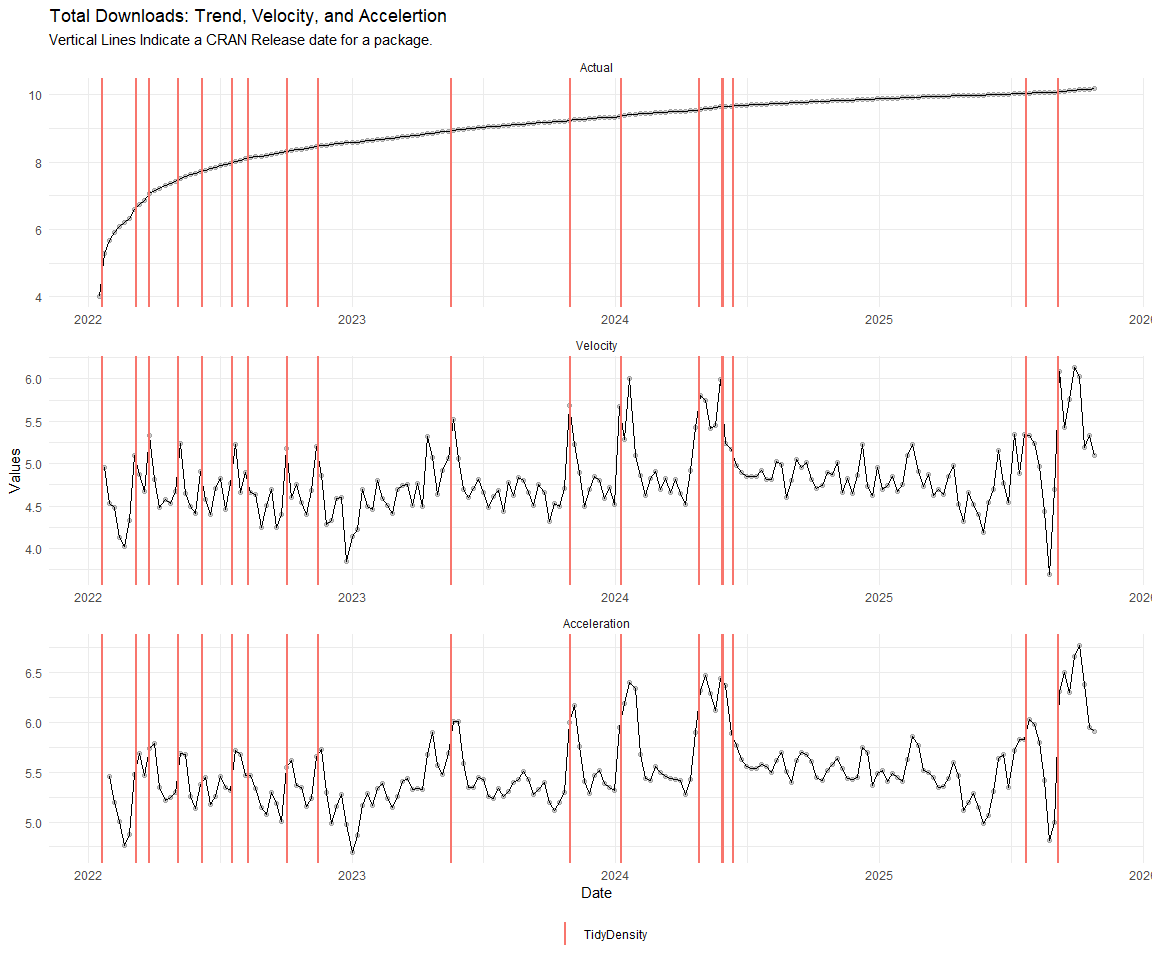
[[3]]
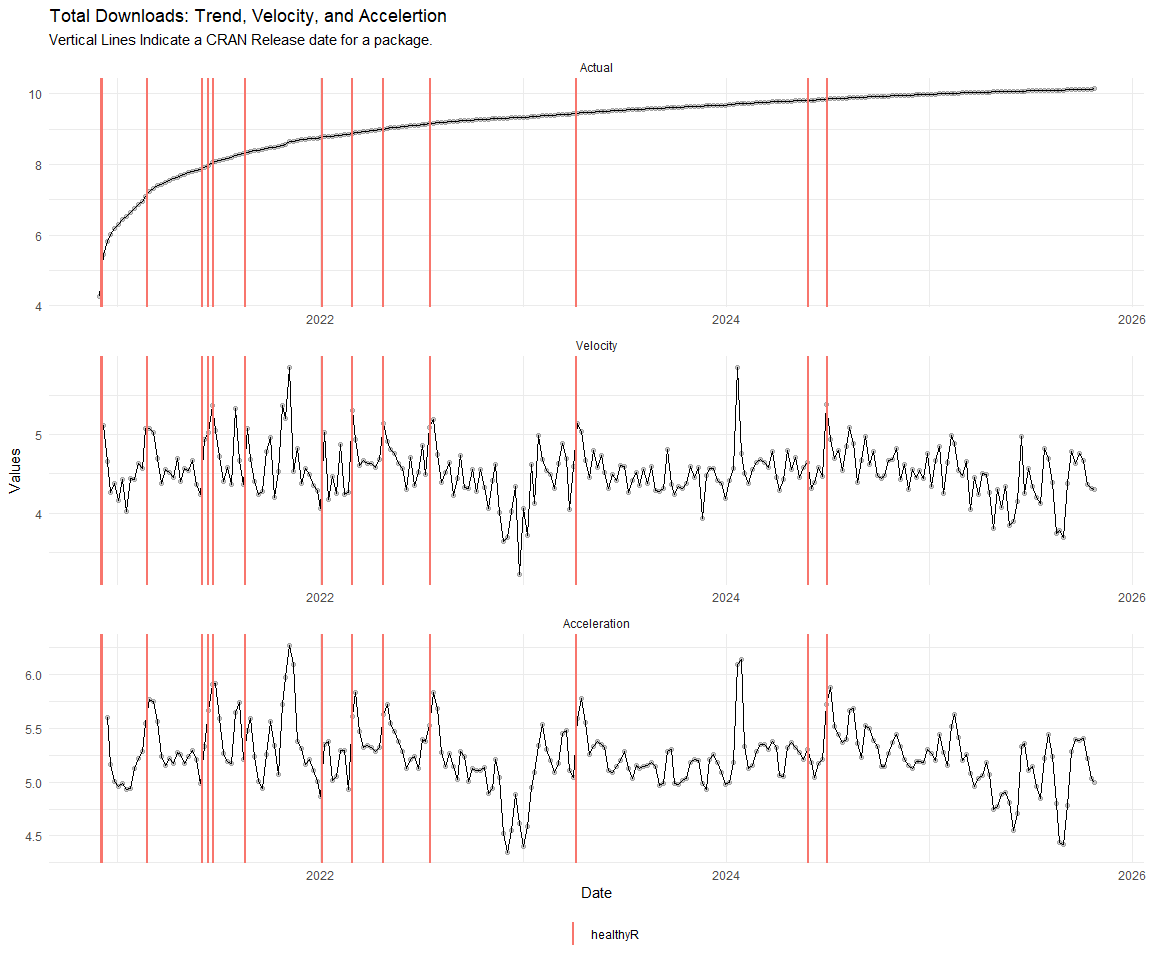
[[4]]
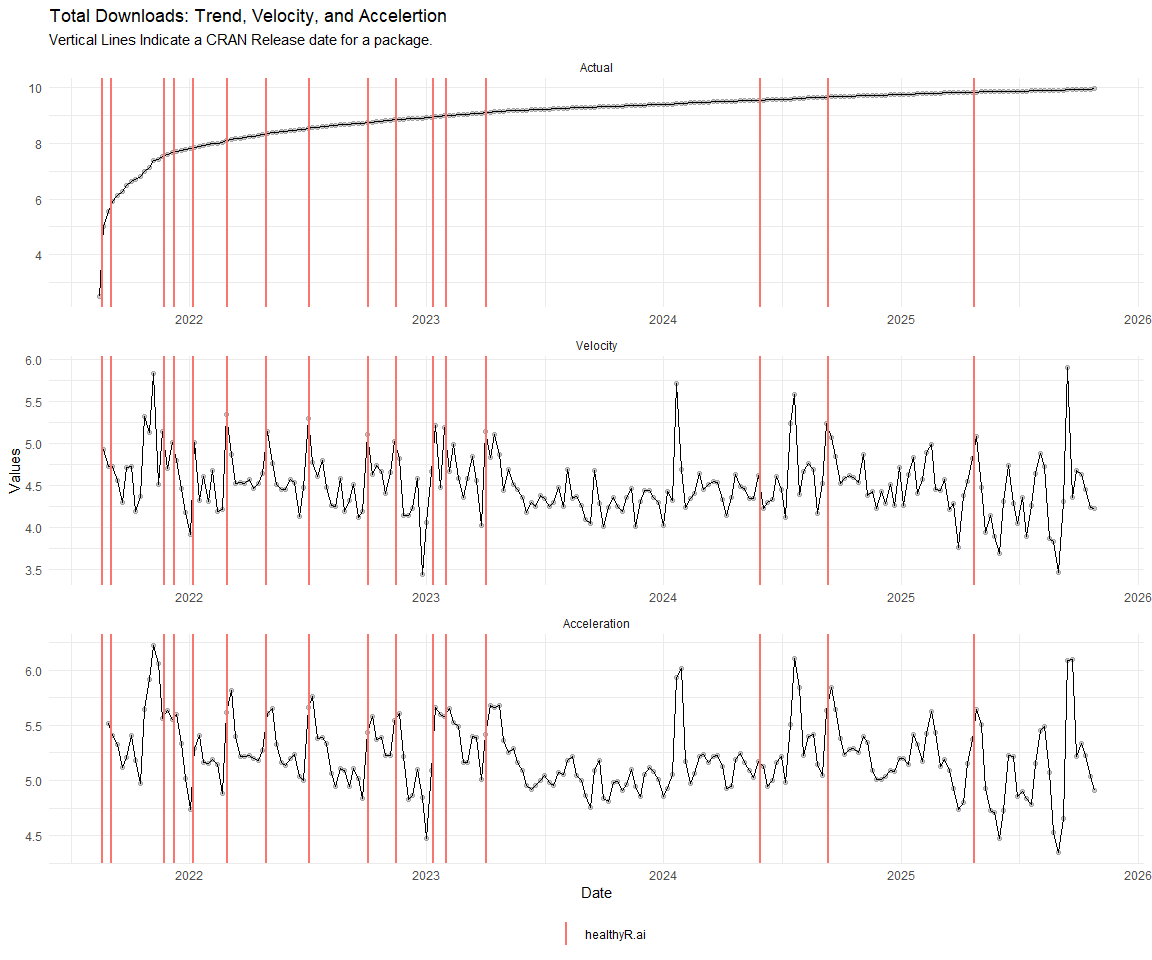
[[5]]
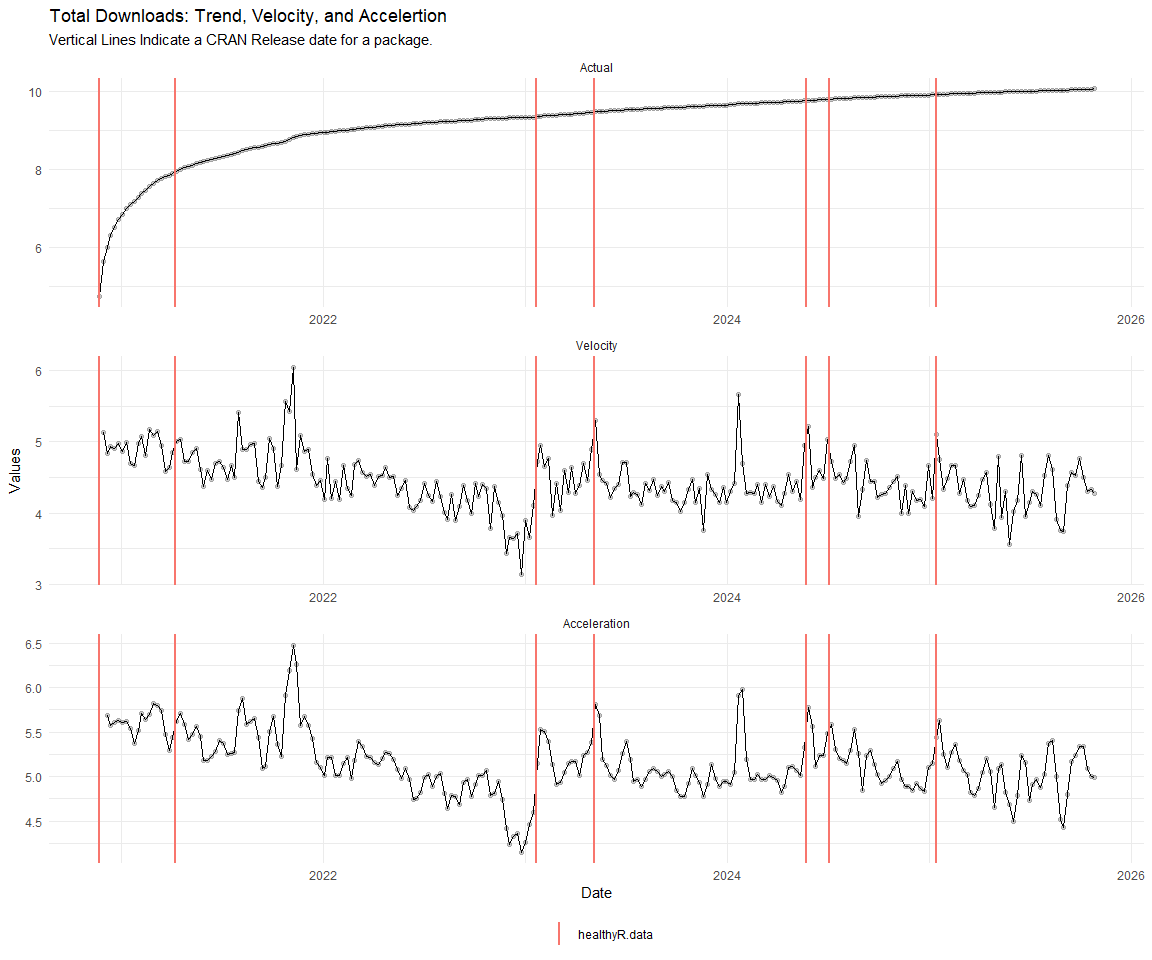
[[6]]
[[7]]
[[8]]
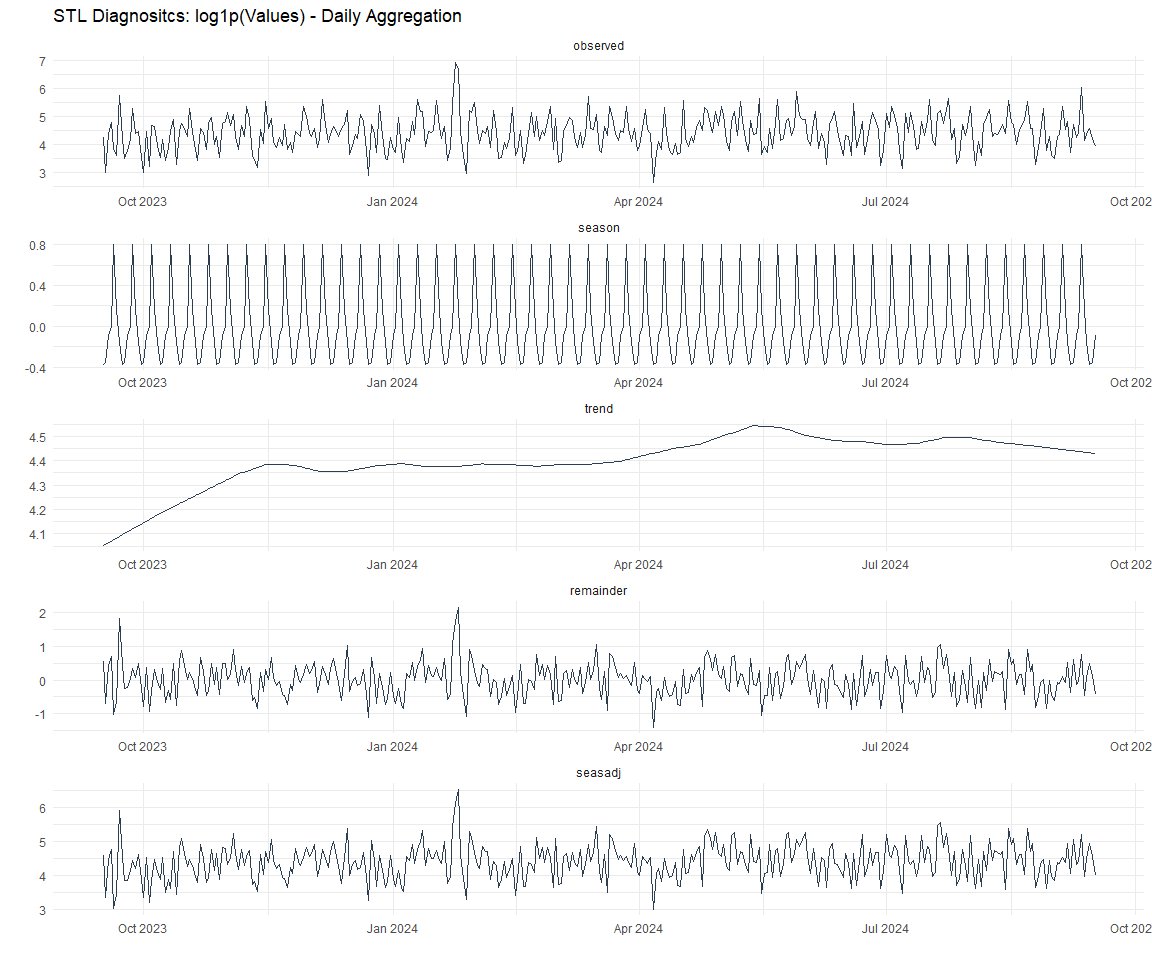
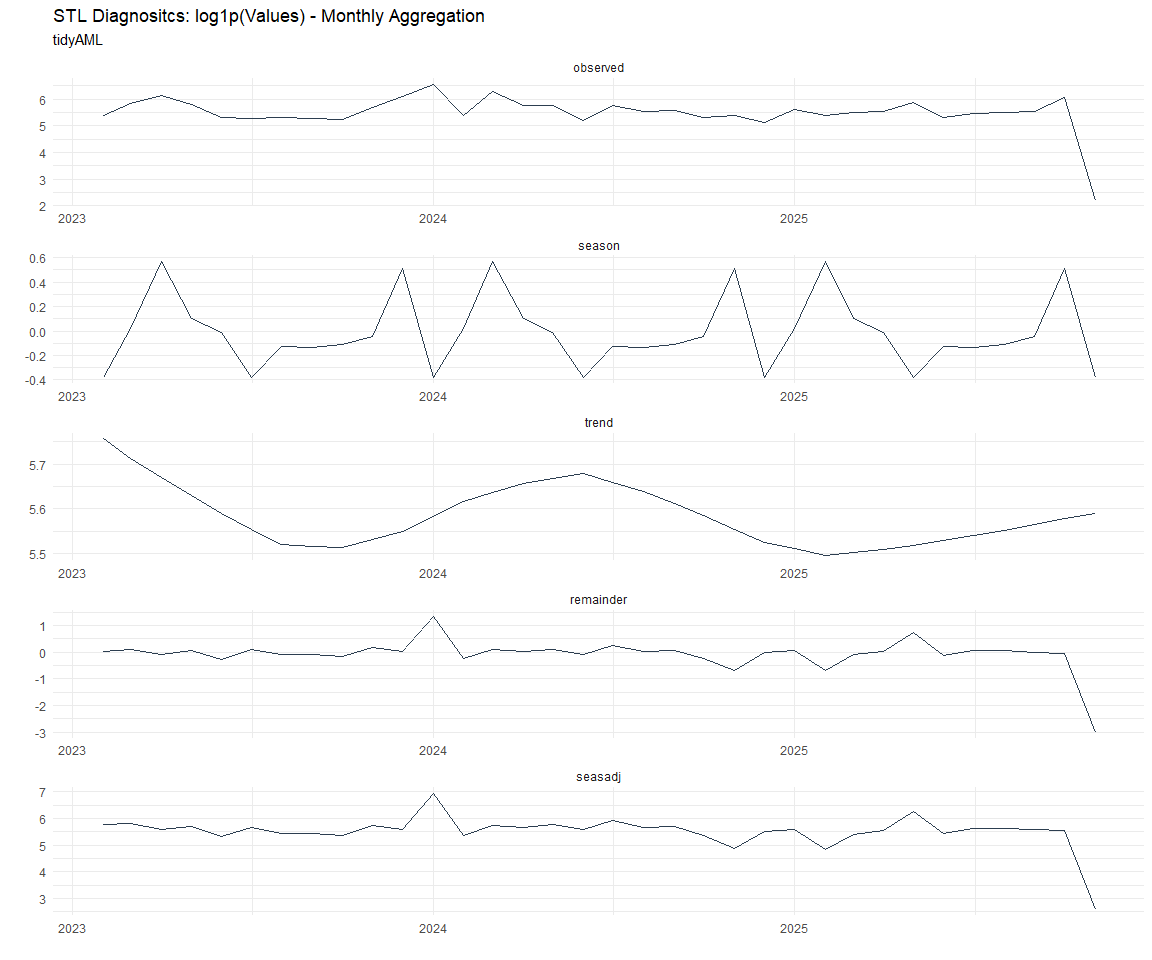
Now lets take a look at some time series decomposition graphs.
[[1]]
[[2]]
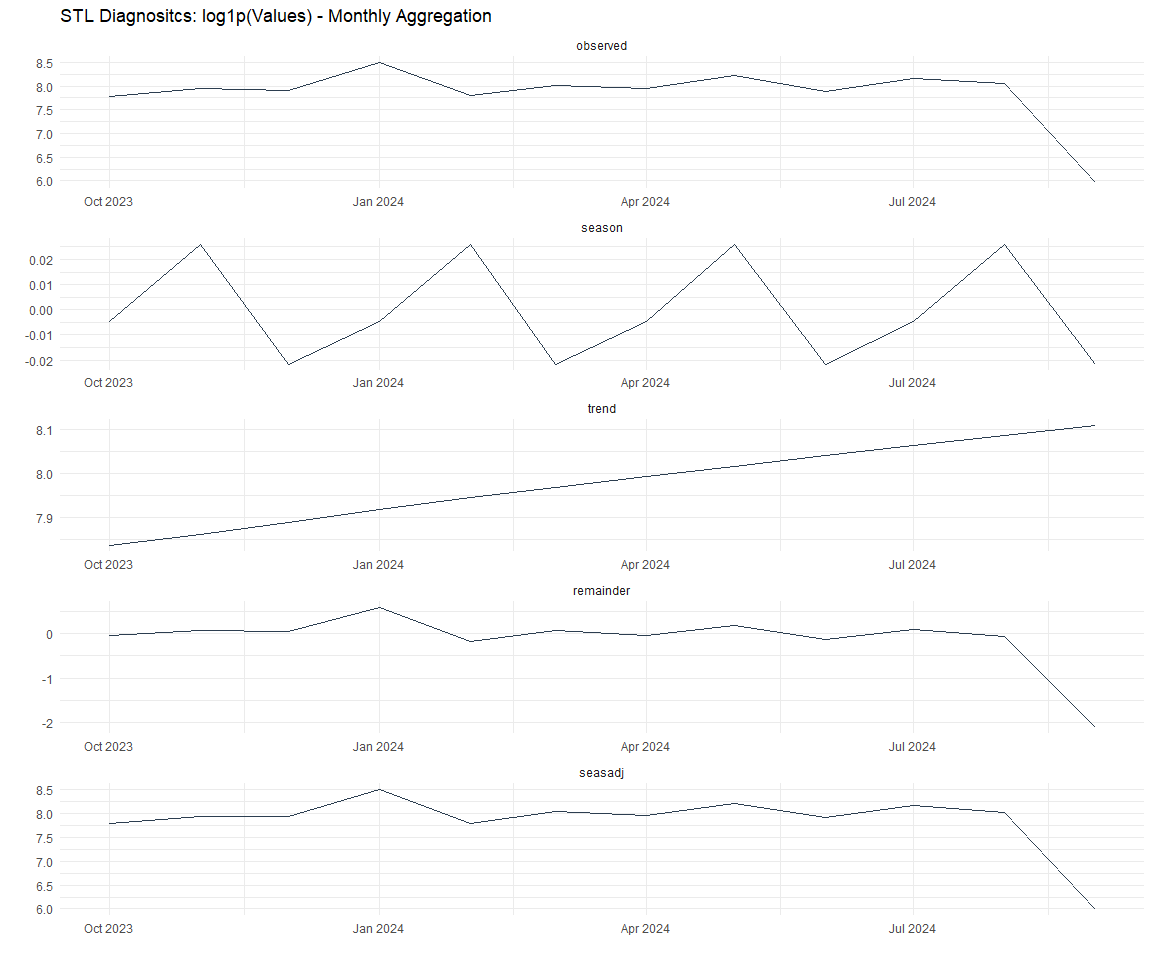
[[3]]
[[4]]

[[5]]
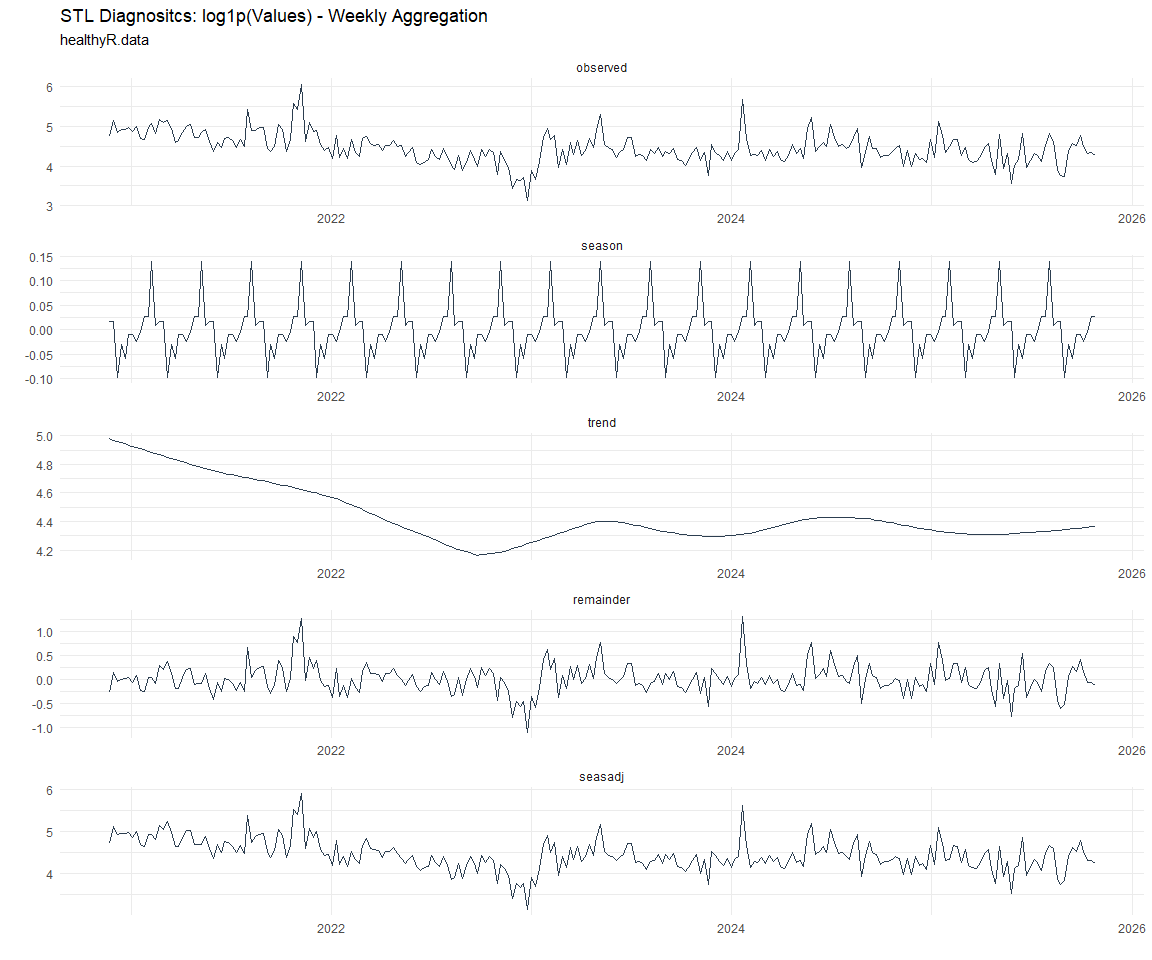
[[6]]
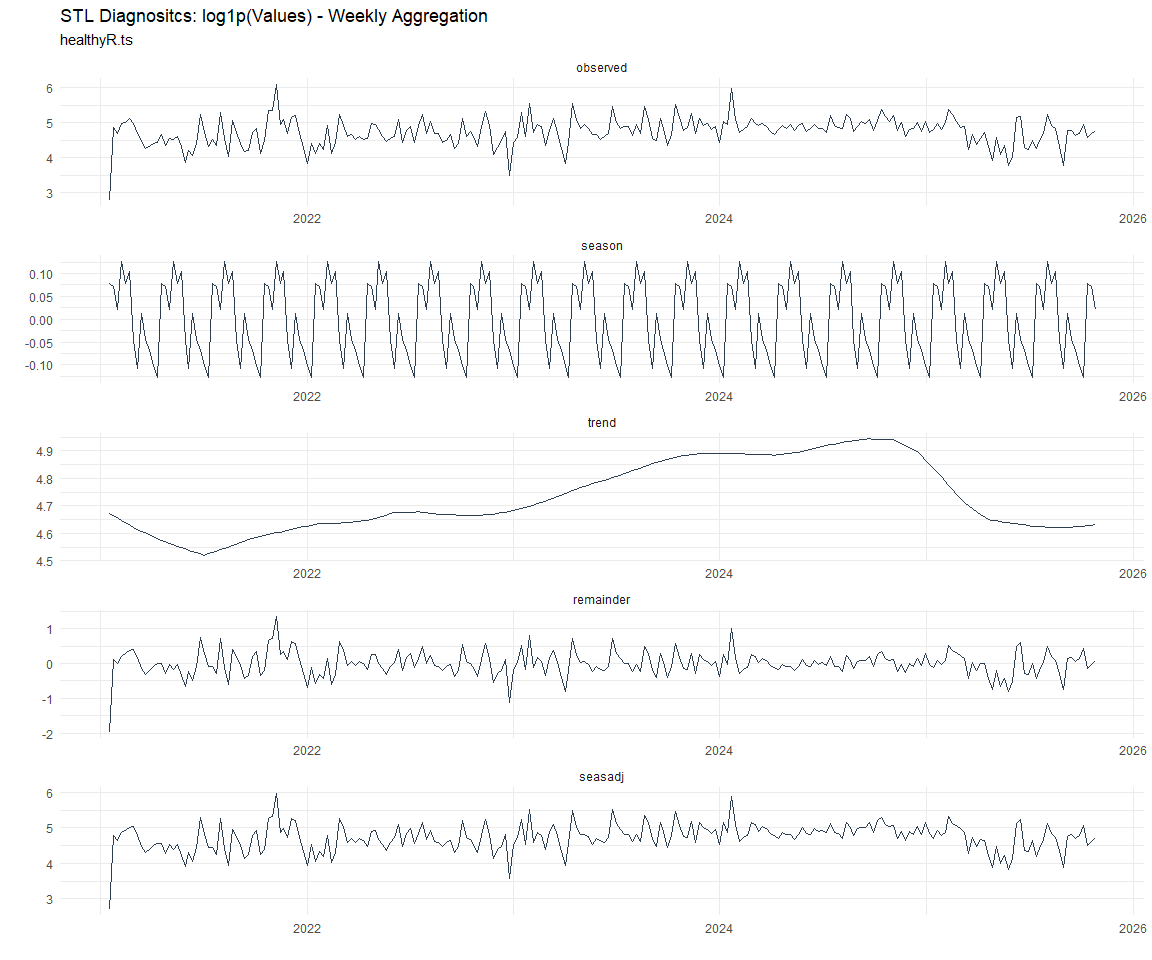
[[7]]
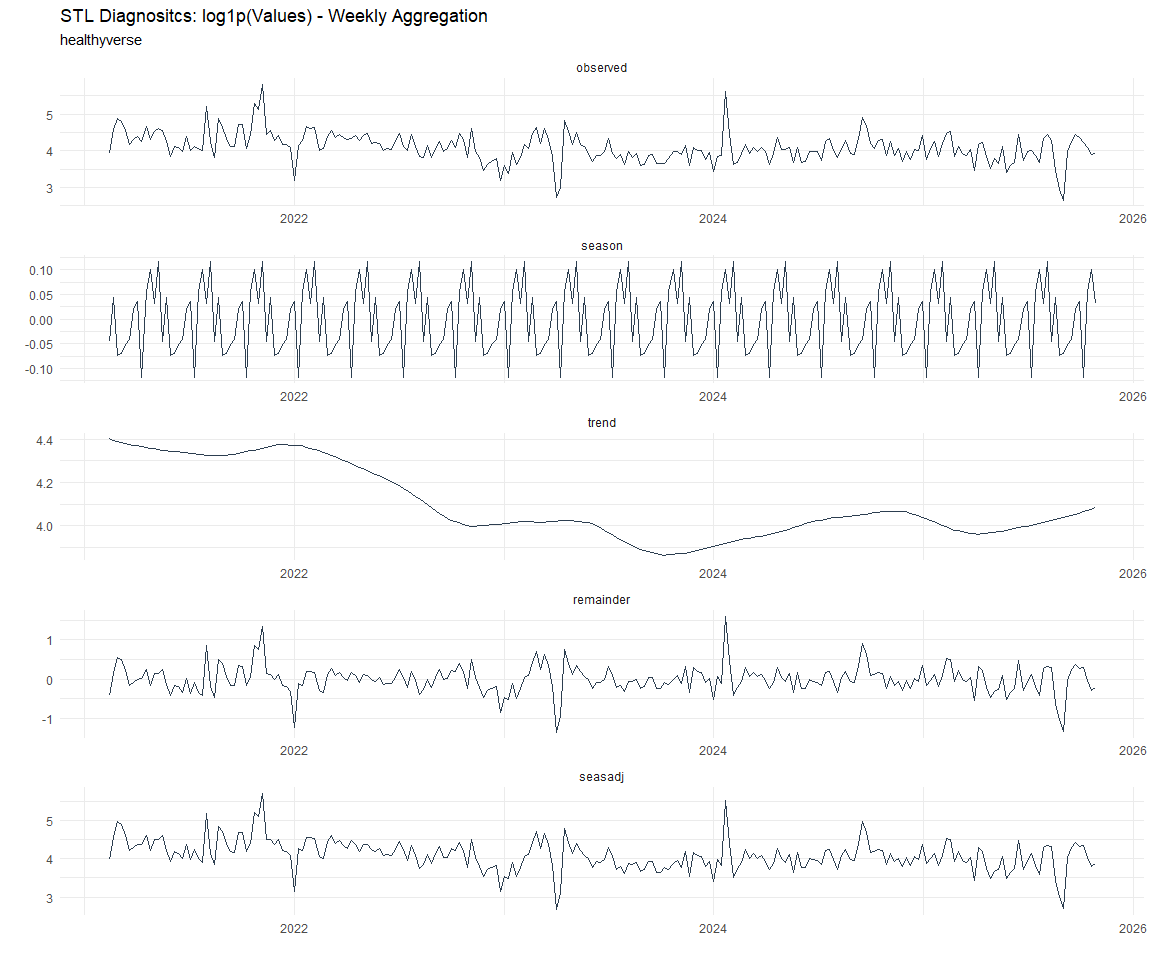
[[8]]
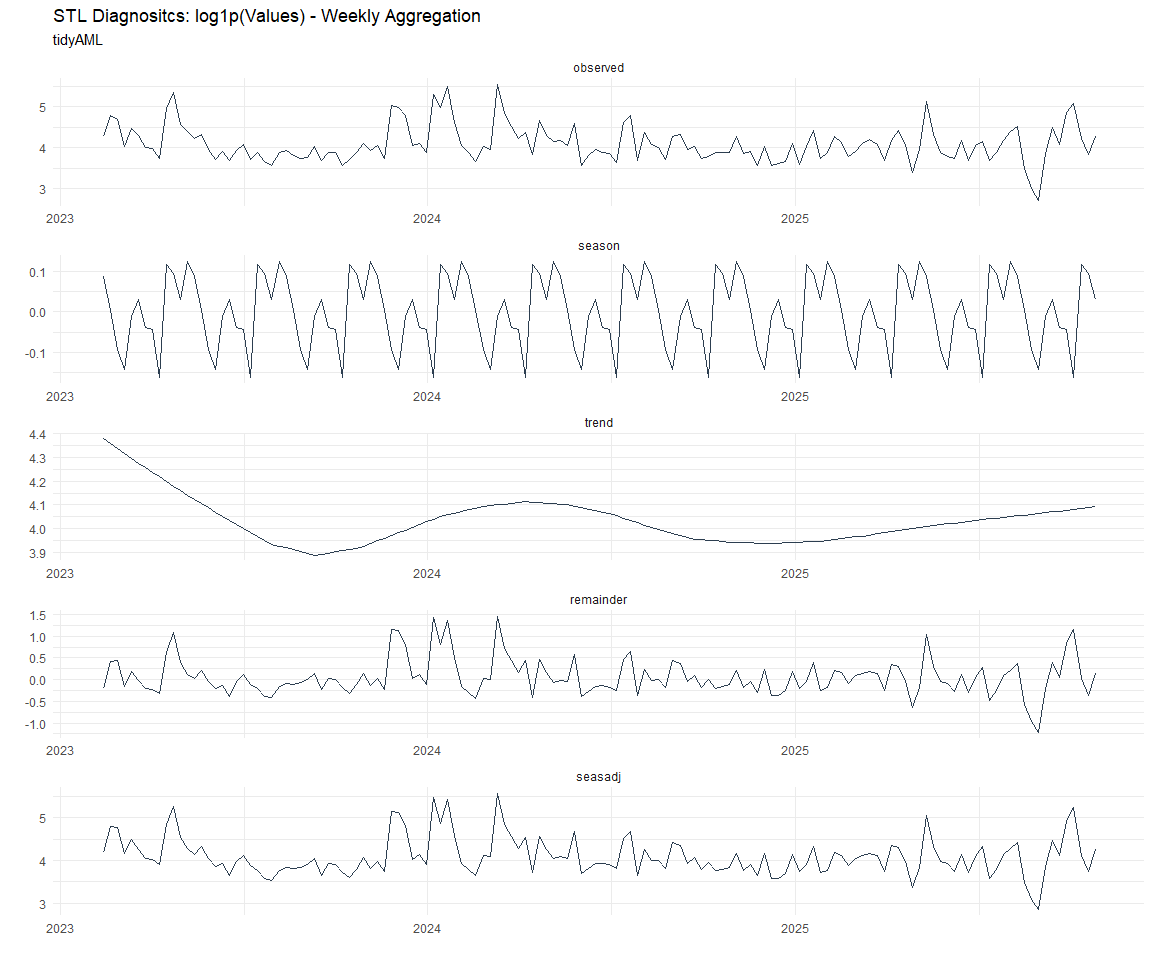
[[1]]
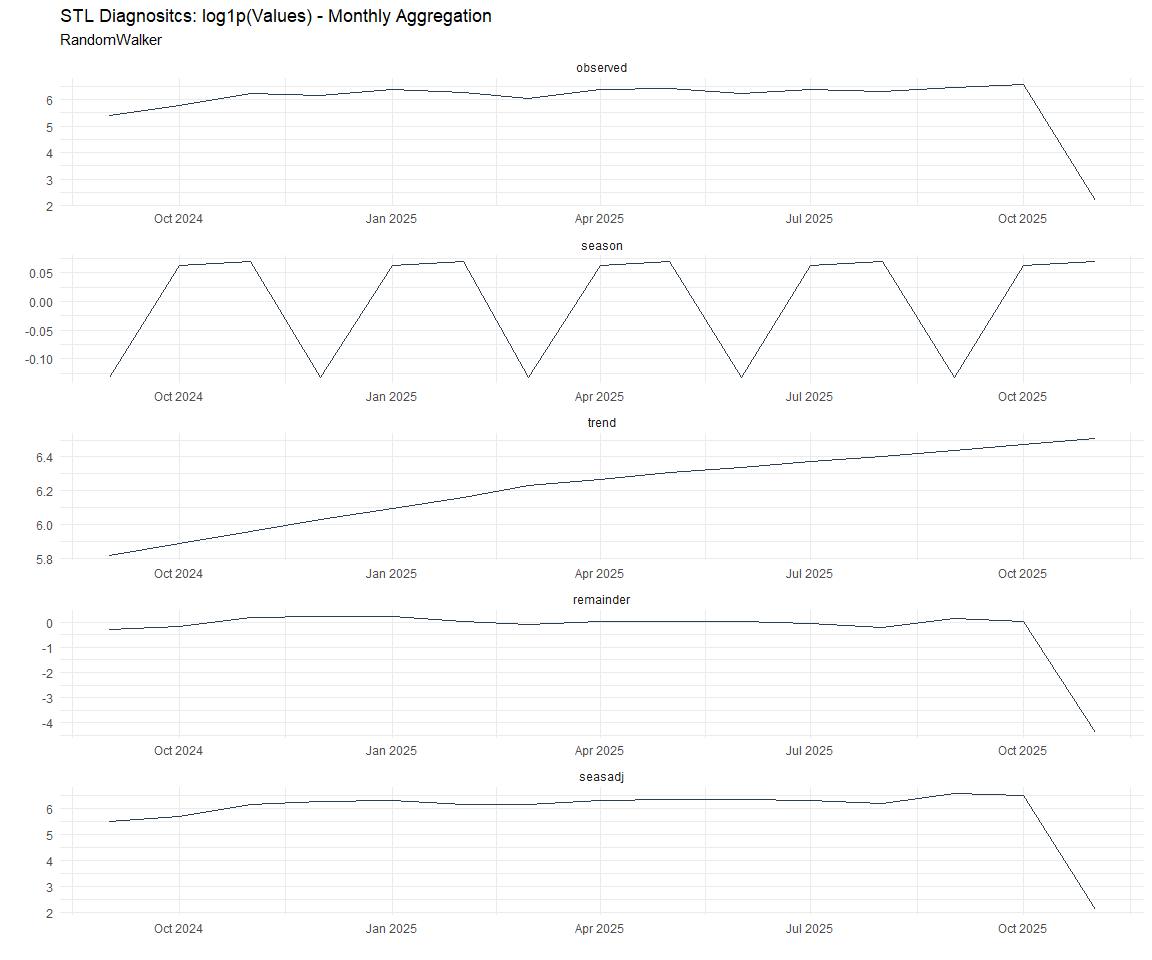
[[2]]
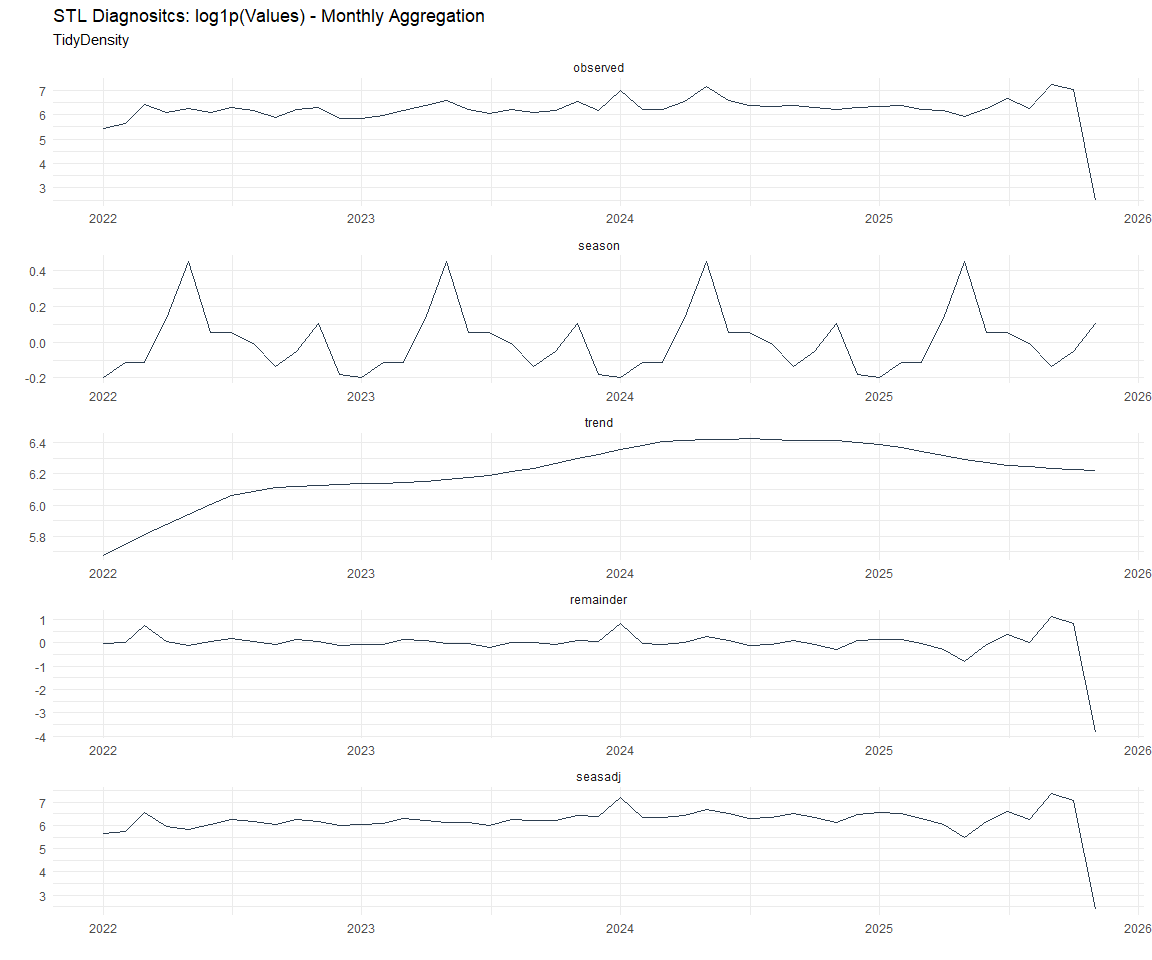
[[3]]
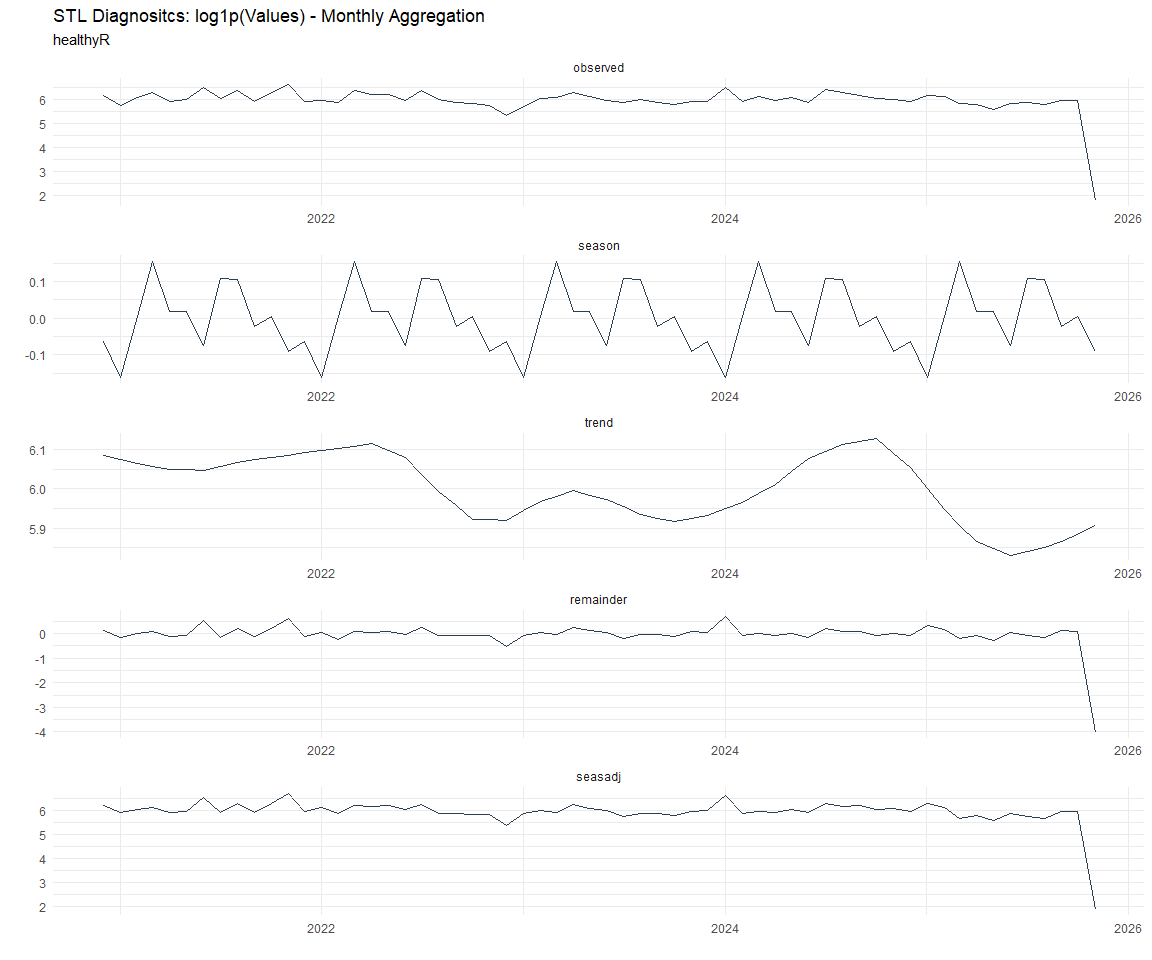
[[4]]
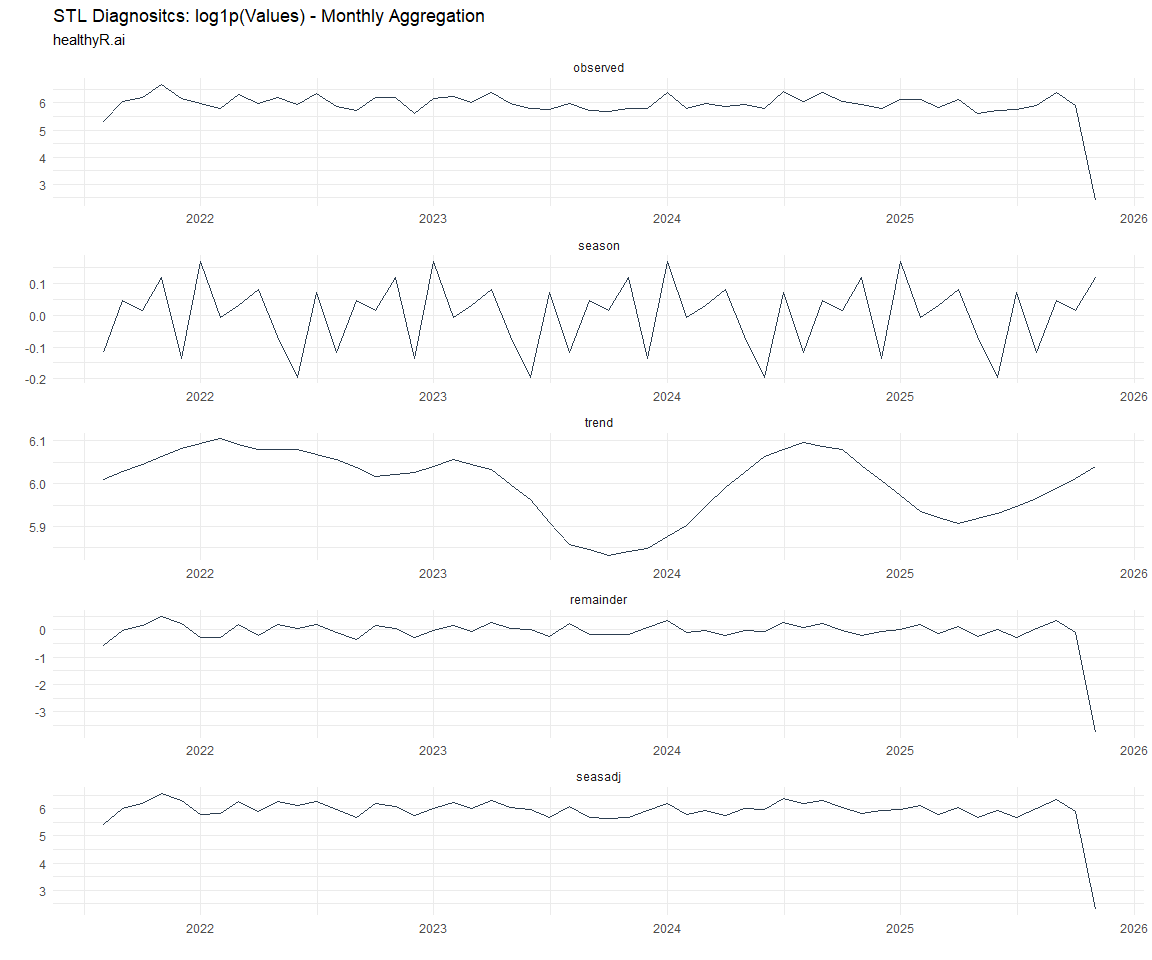
[[5]]
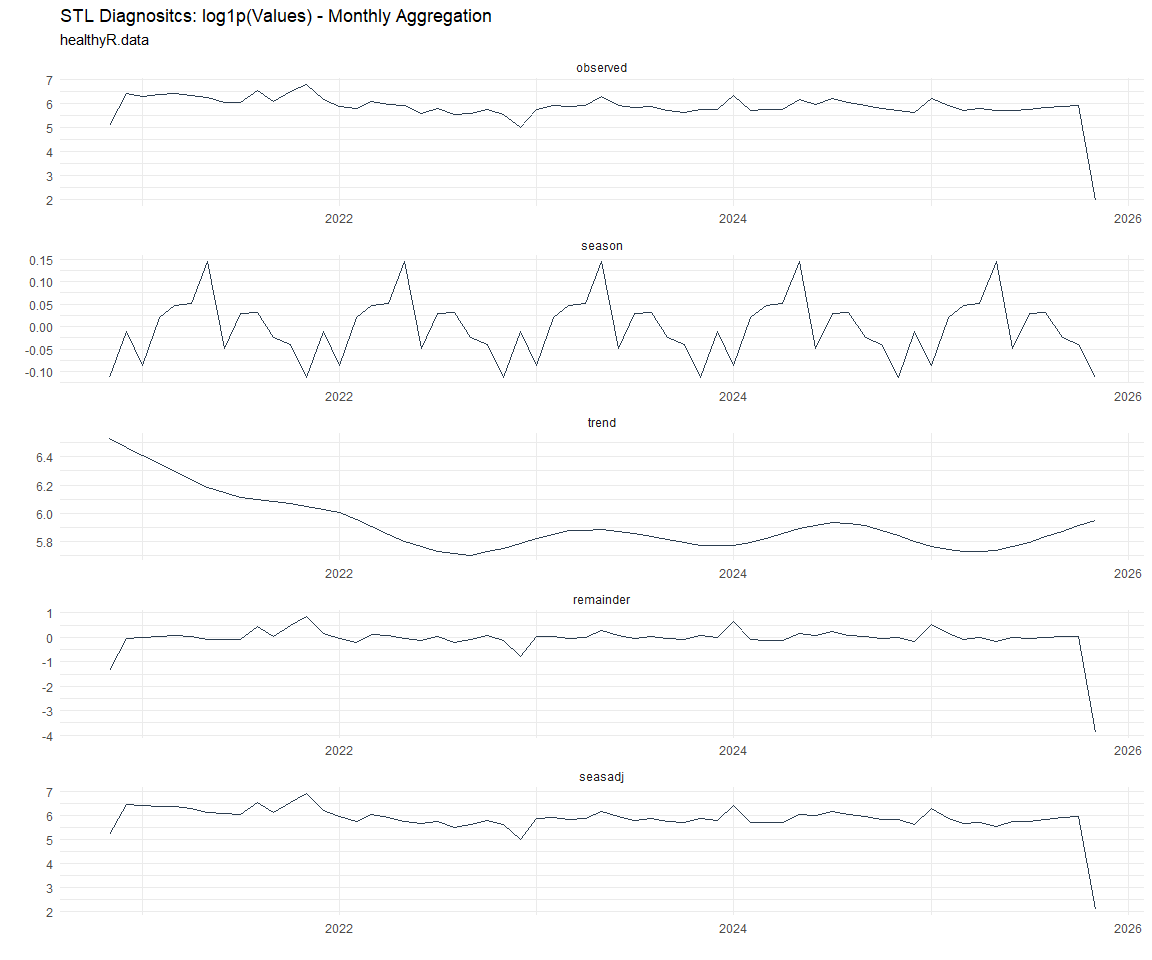
[[6]]
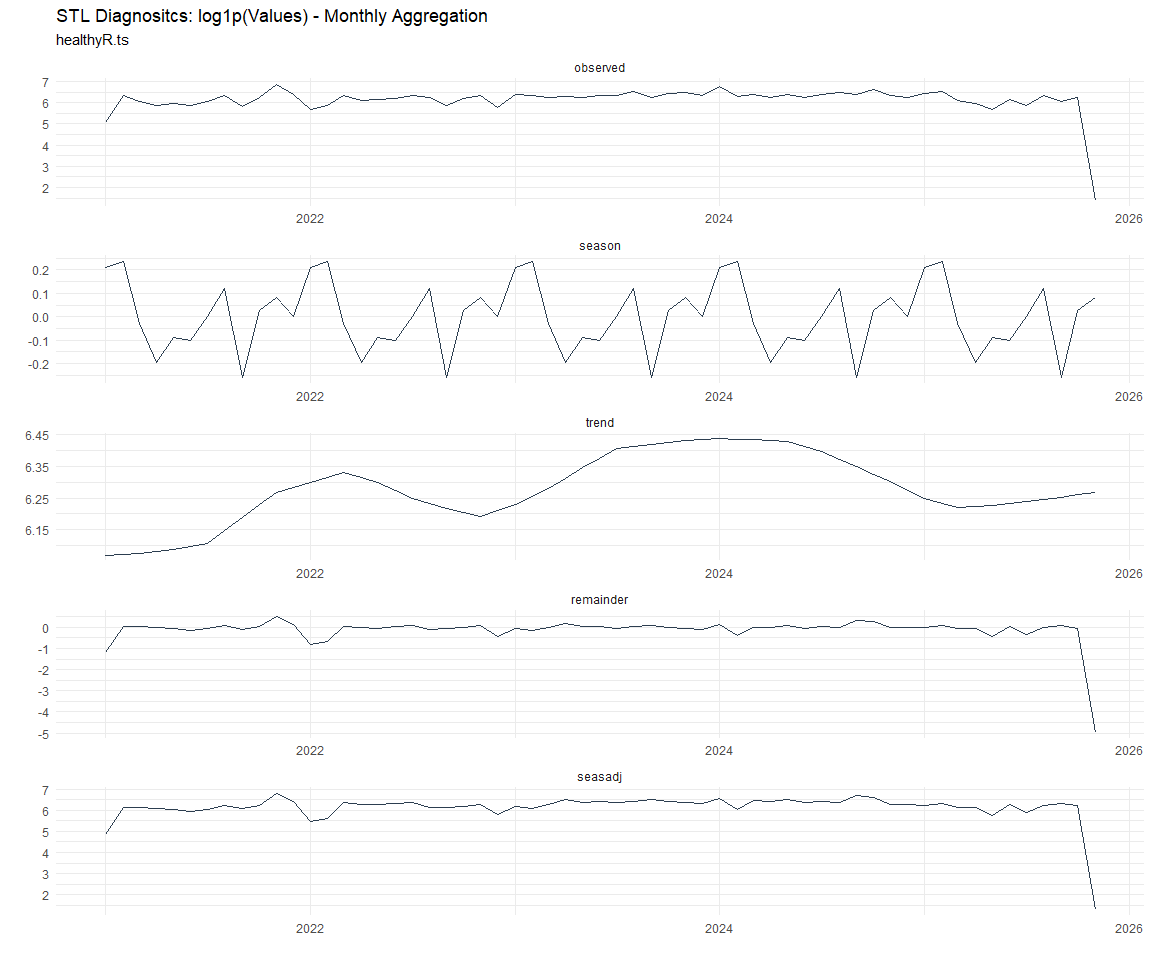
[[7]]
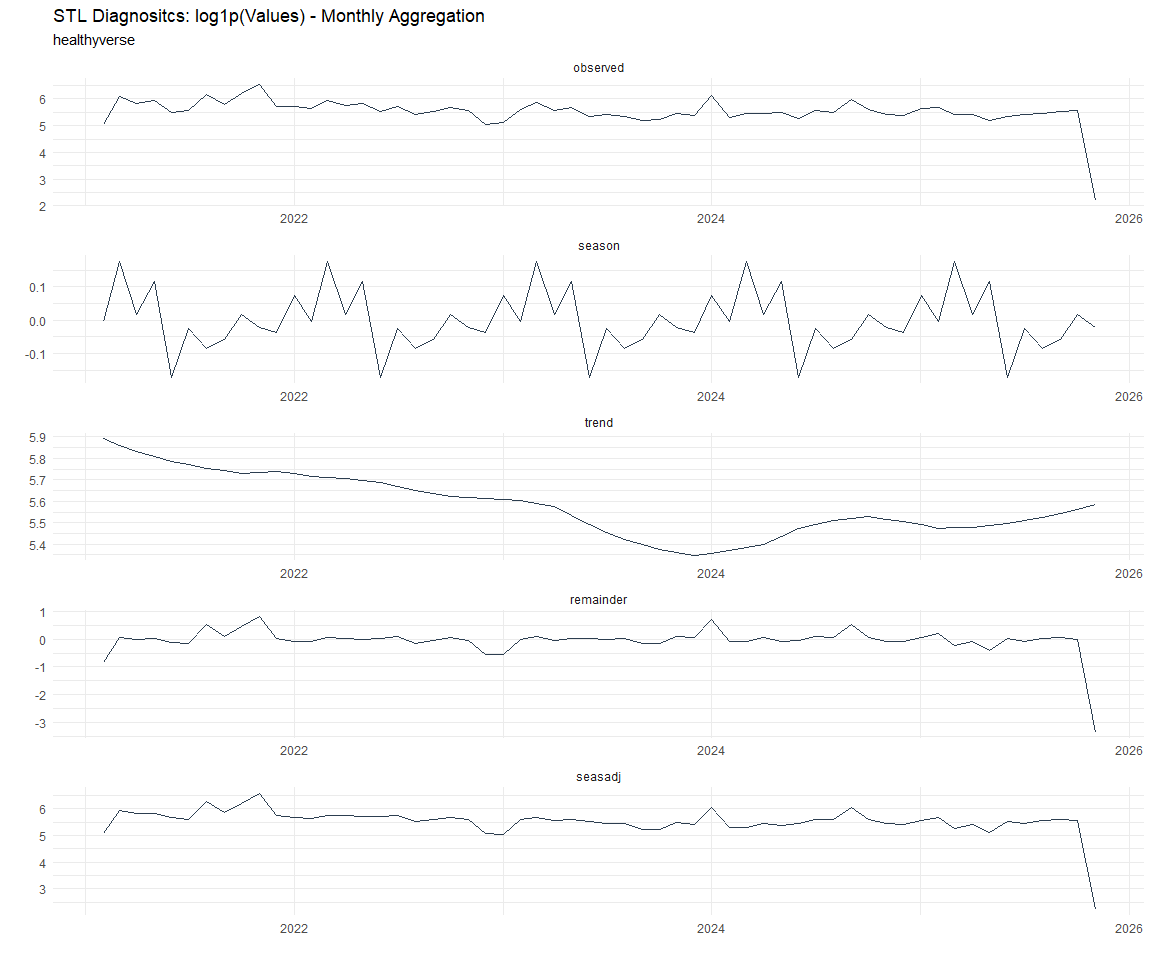
[[8]]
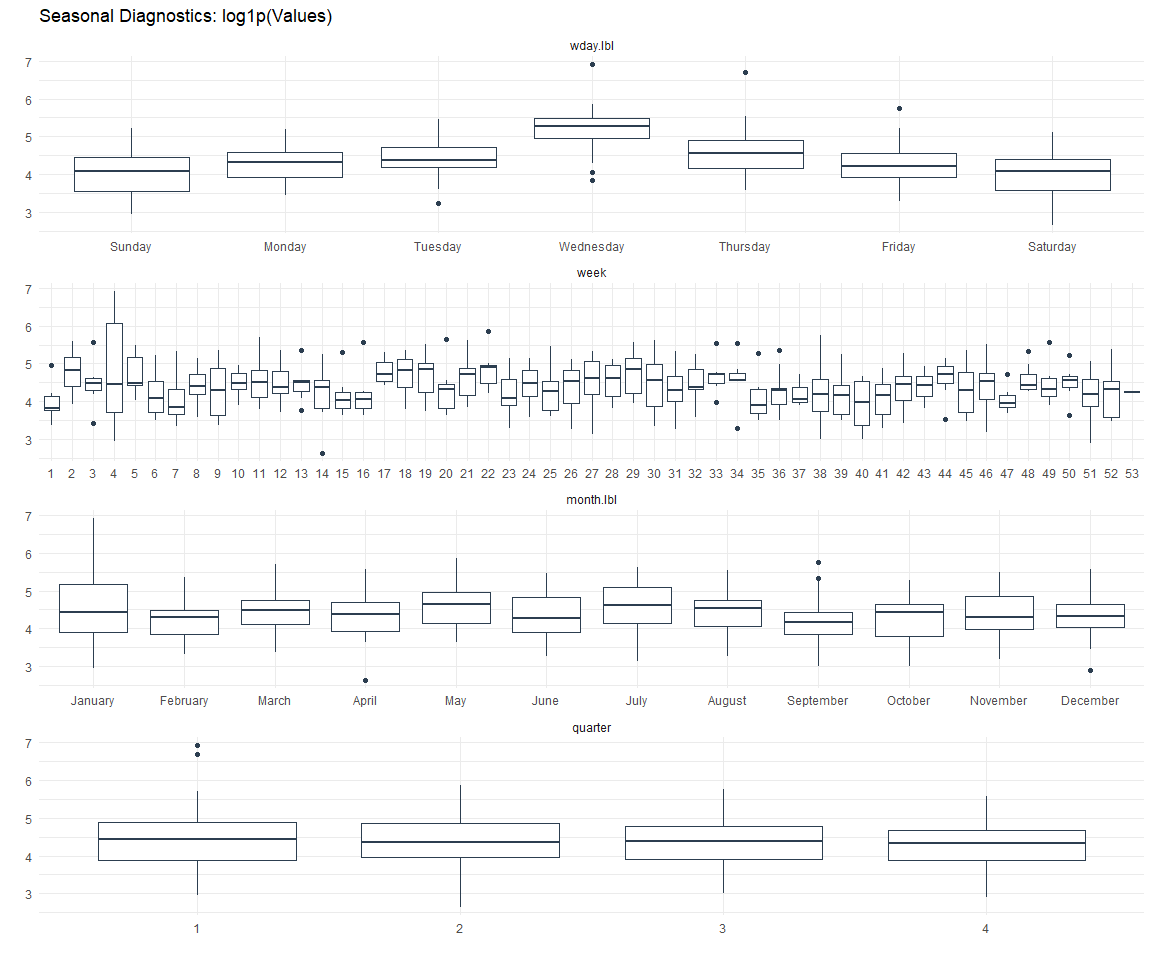
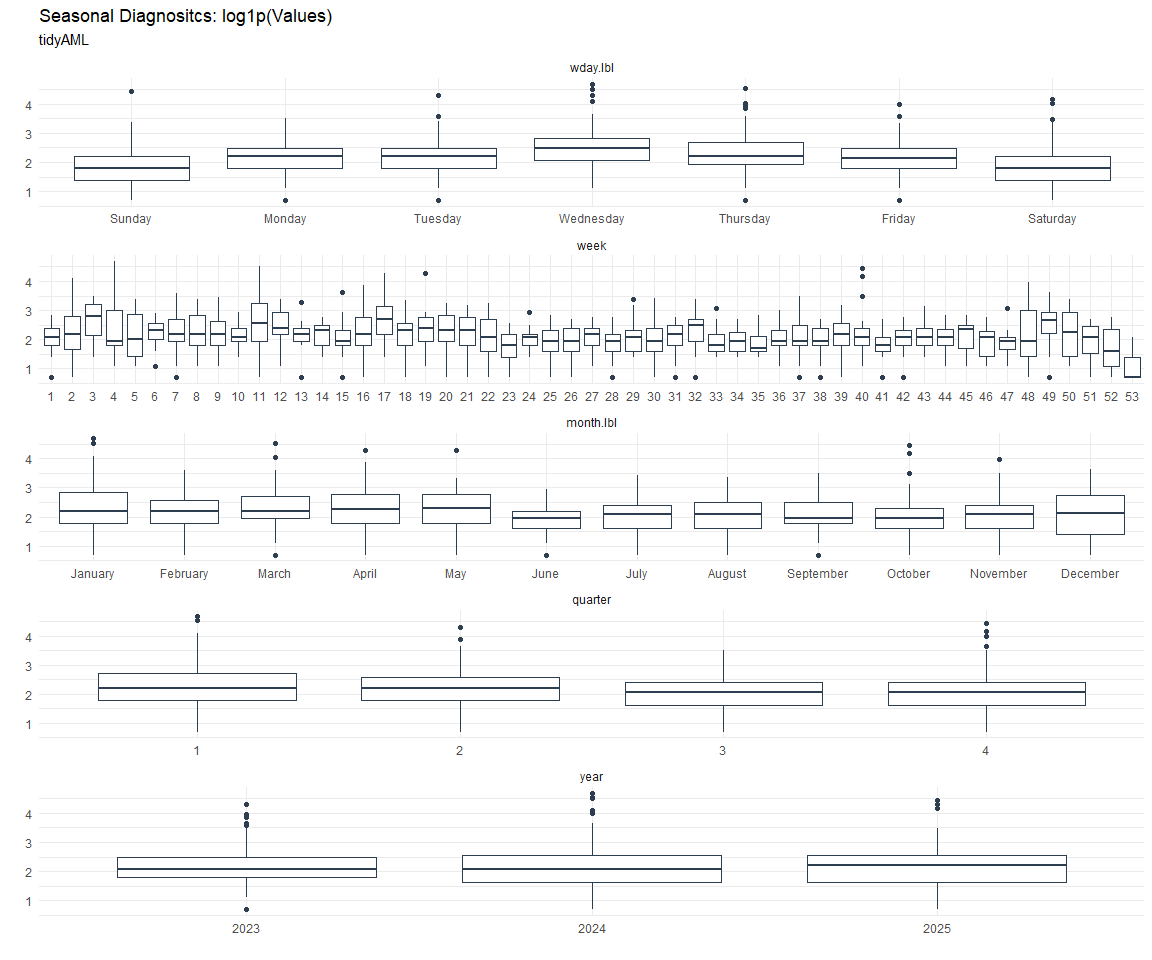
Seasonal Diagnostics:
[[1]]
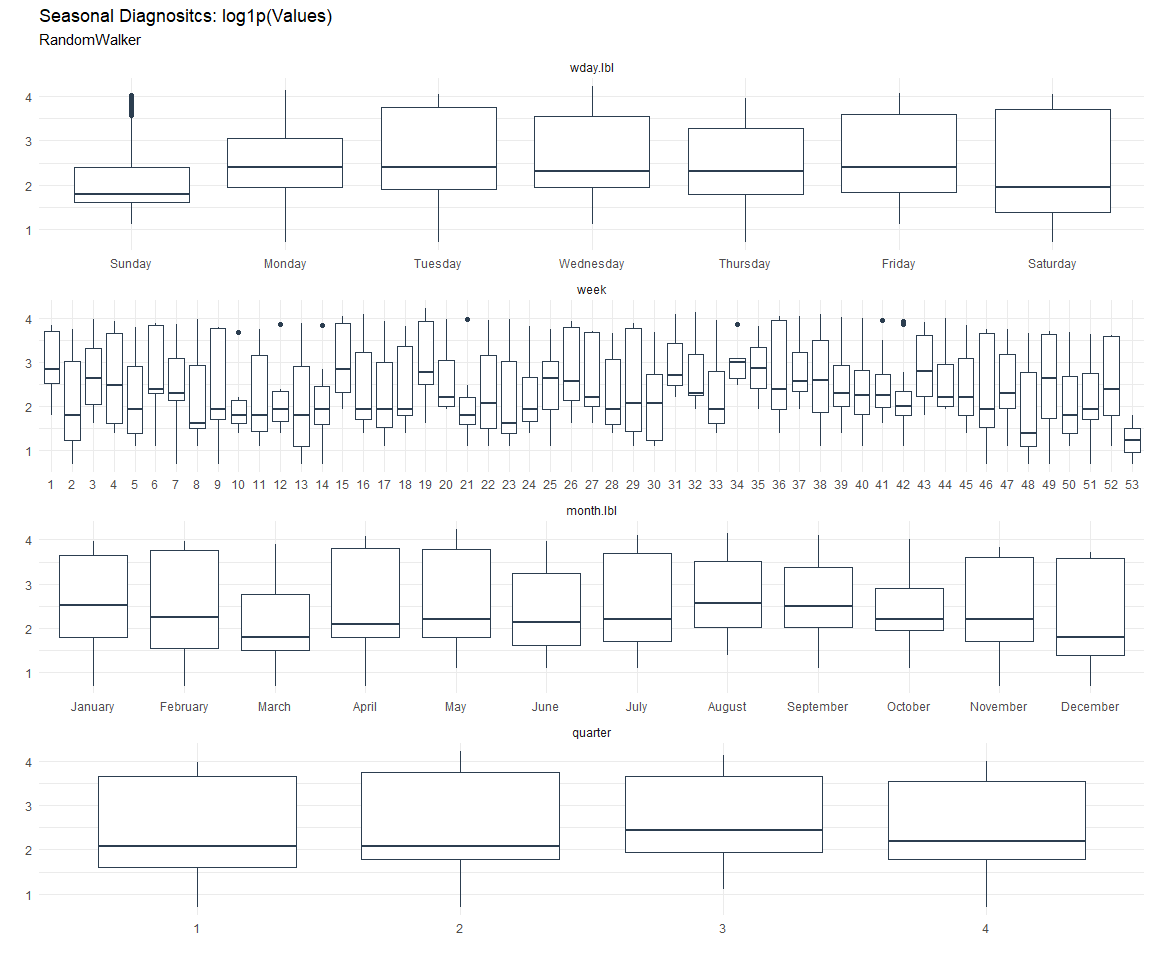
[[2]]
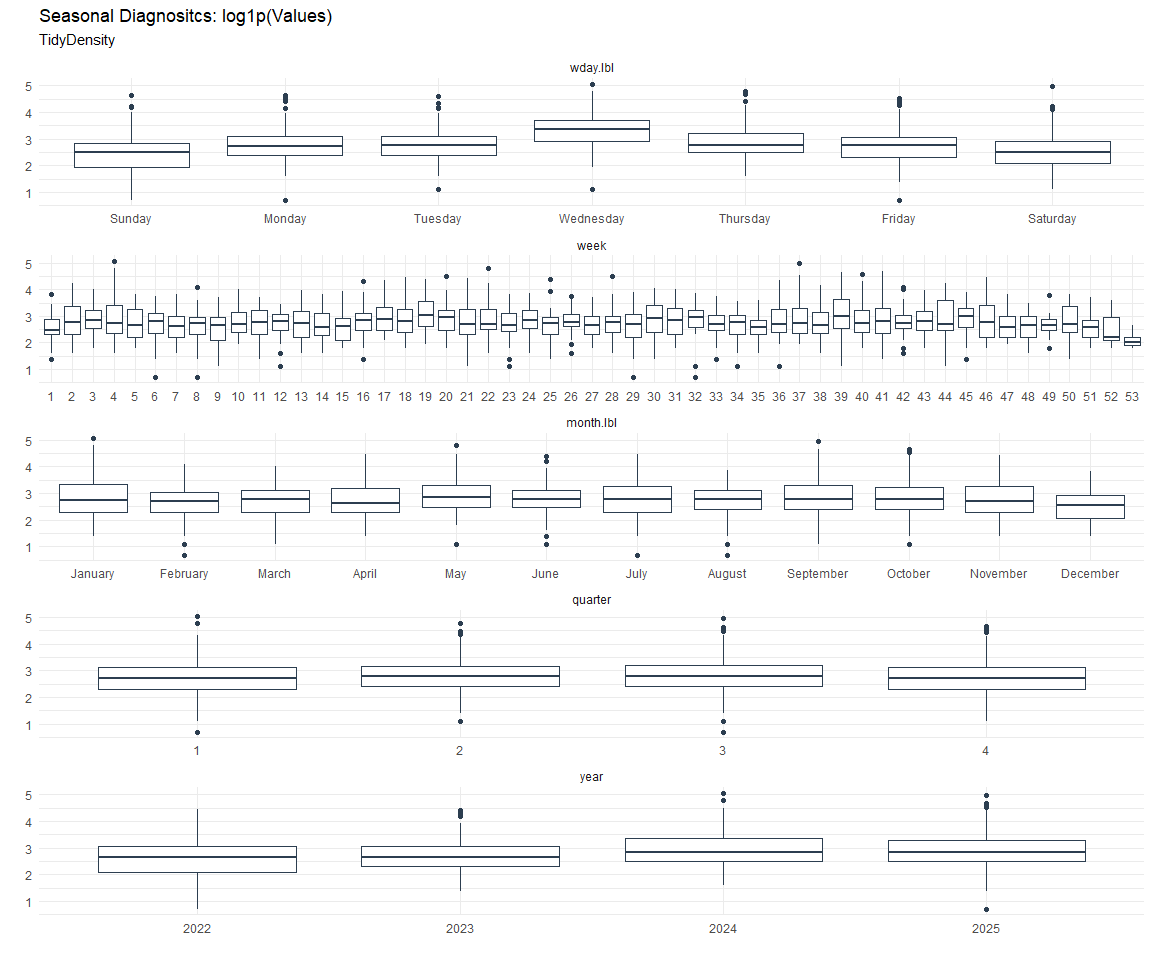
[[3]]
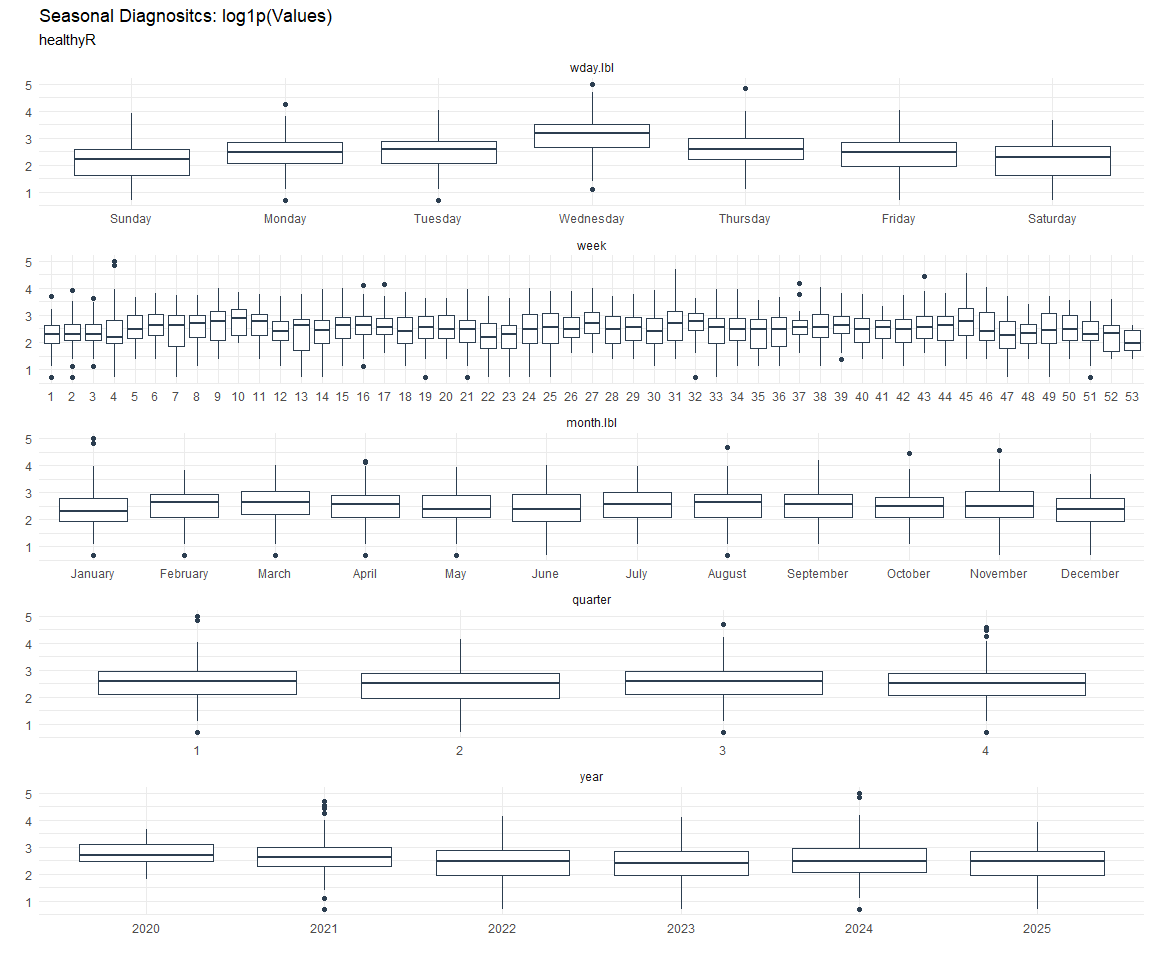
[[4]]
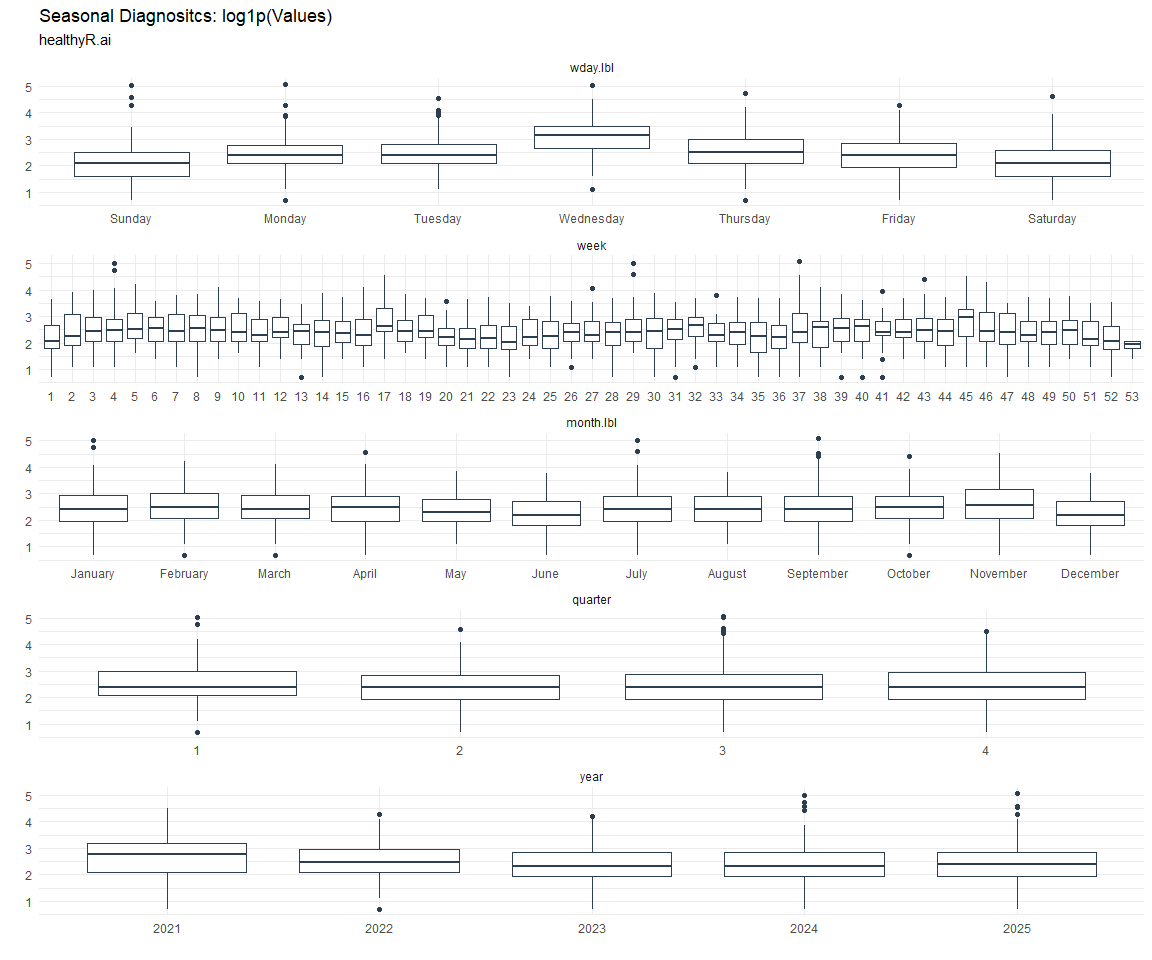
[[5]]
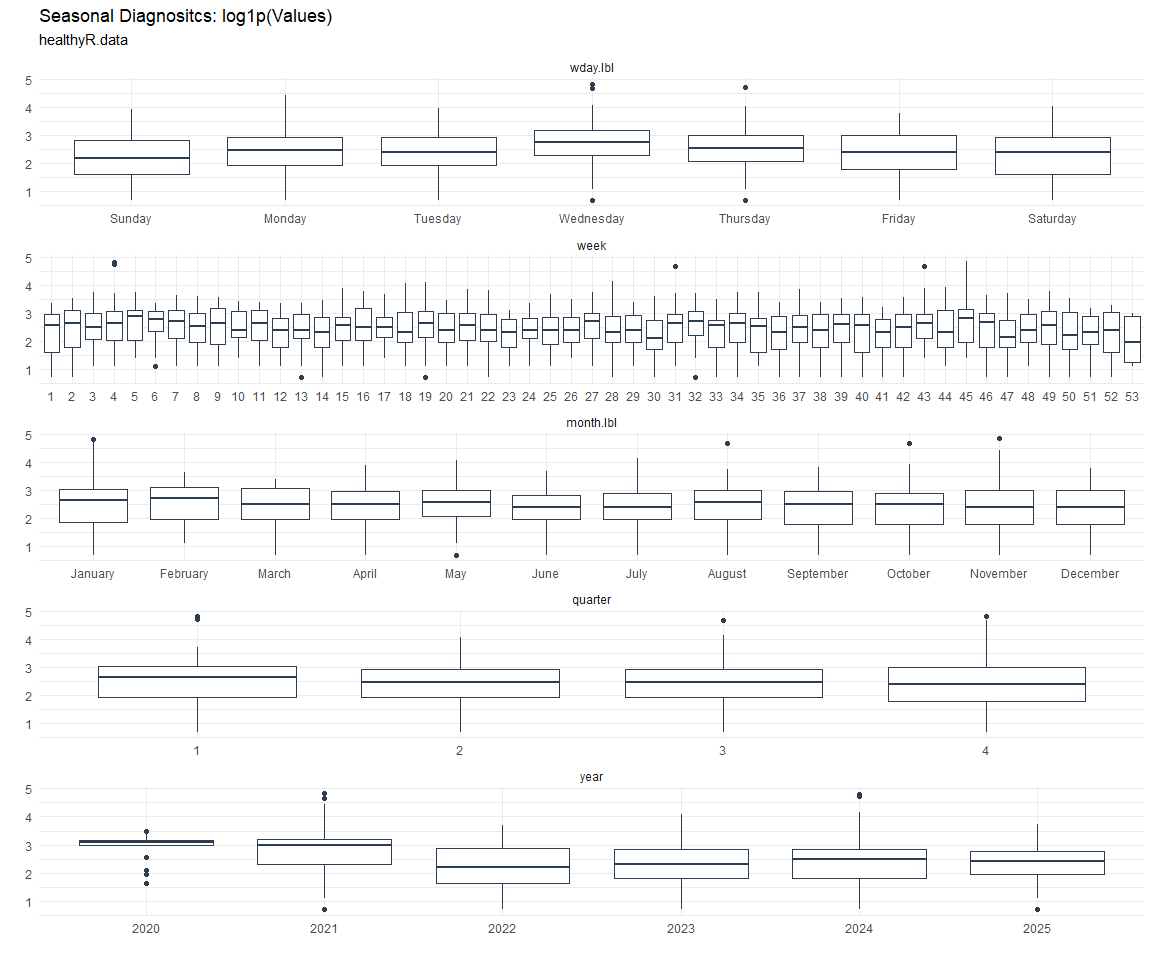
[[6]]
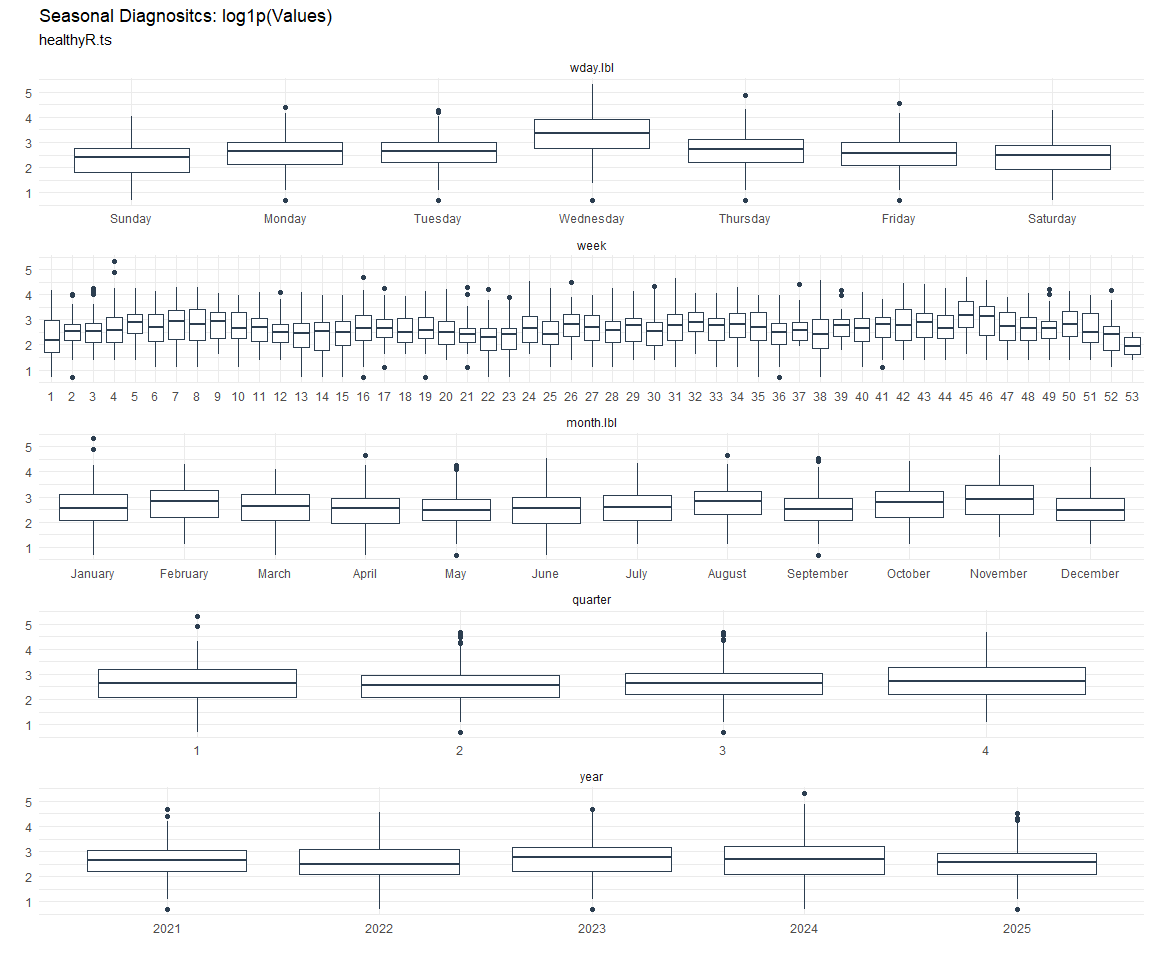
[[7]]
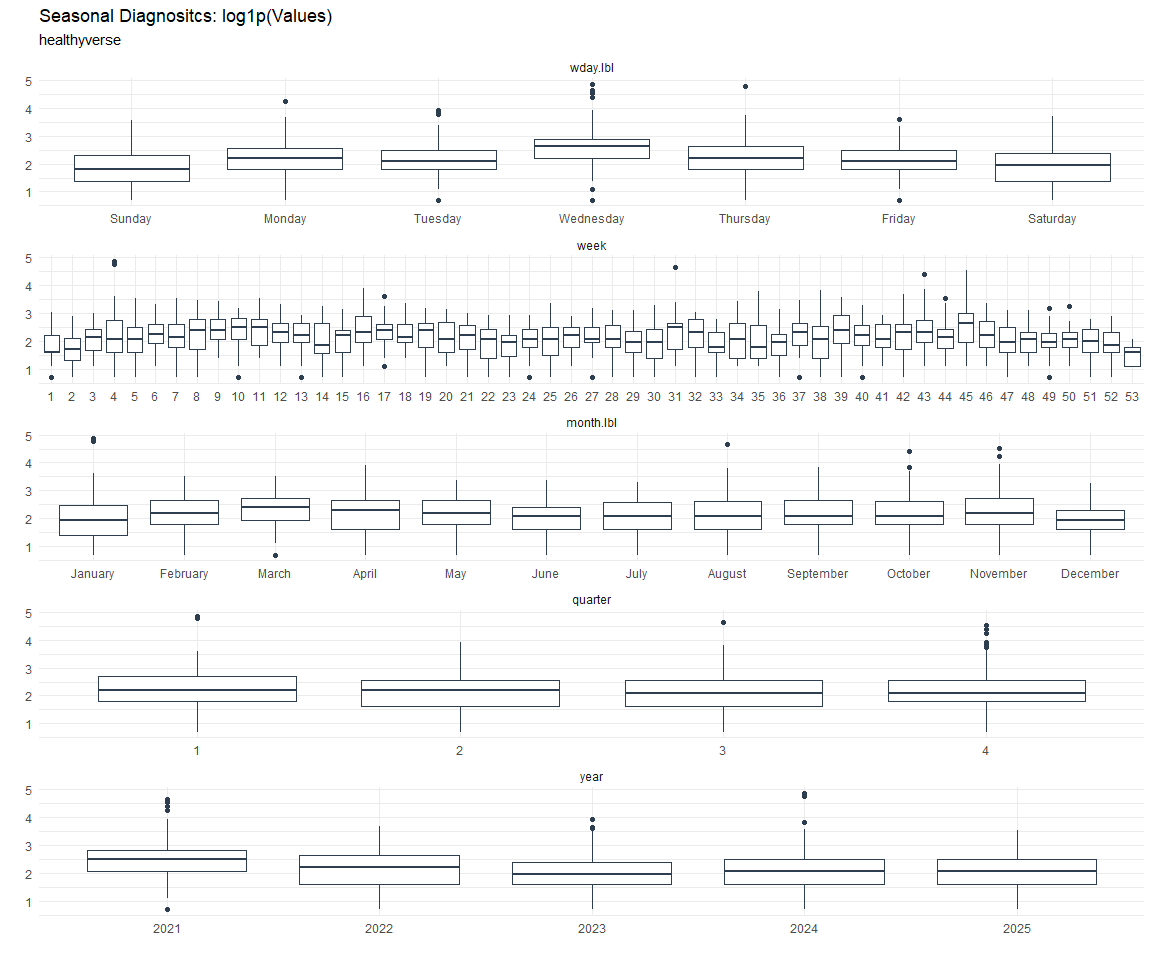
[[8]]
ACF and PACF Diagnostics:
[[1]]
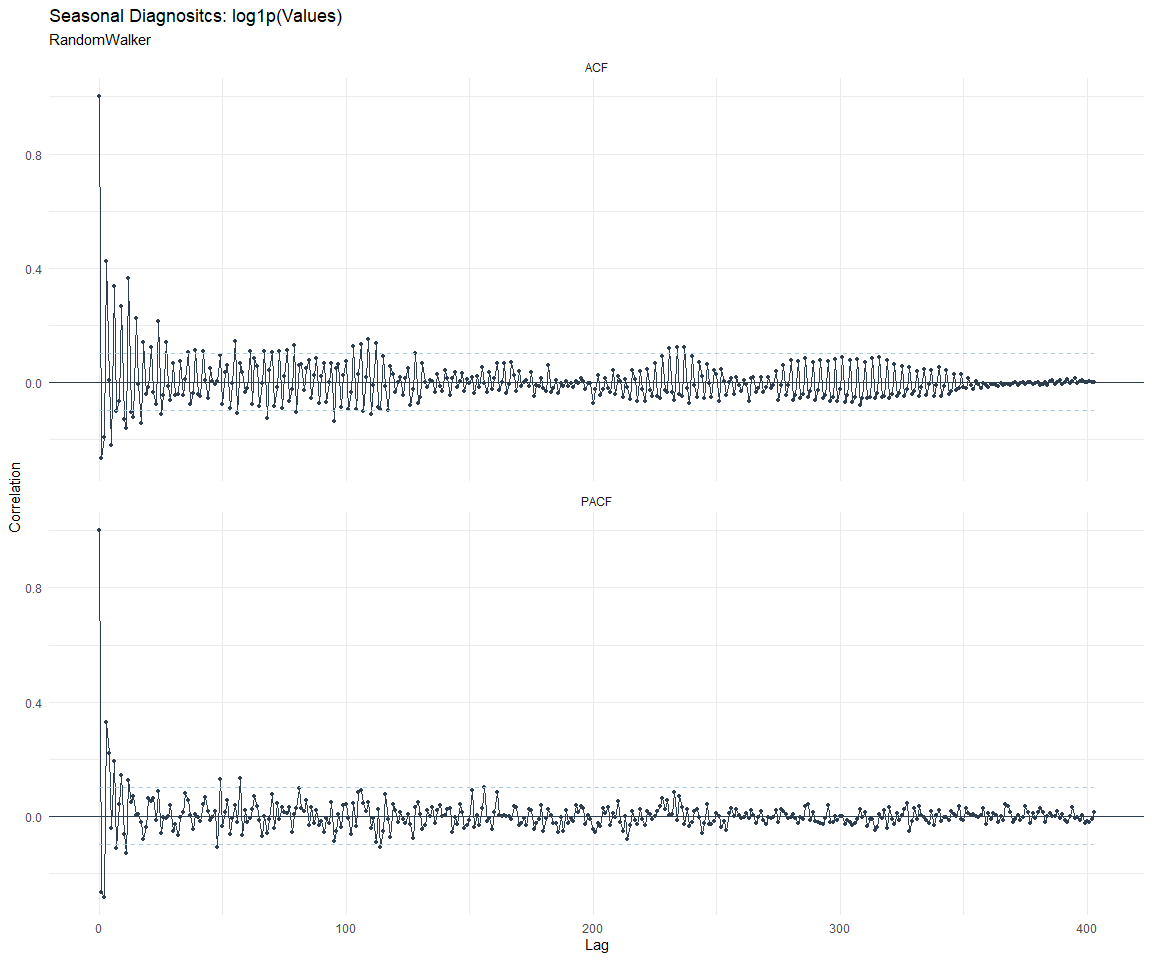
[[2]]
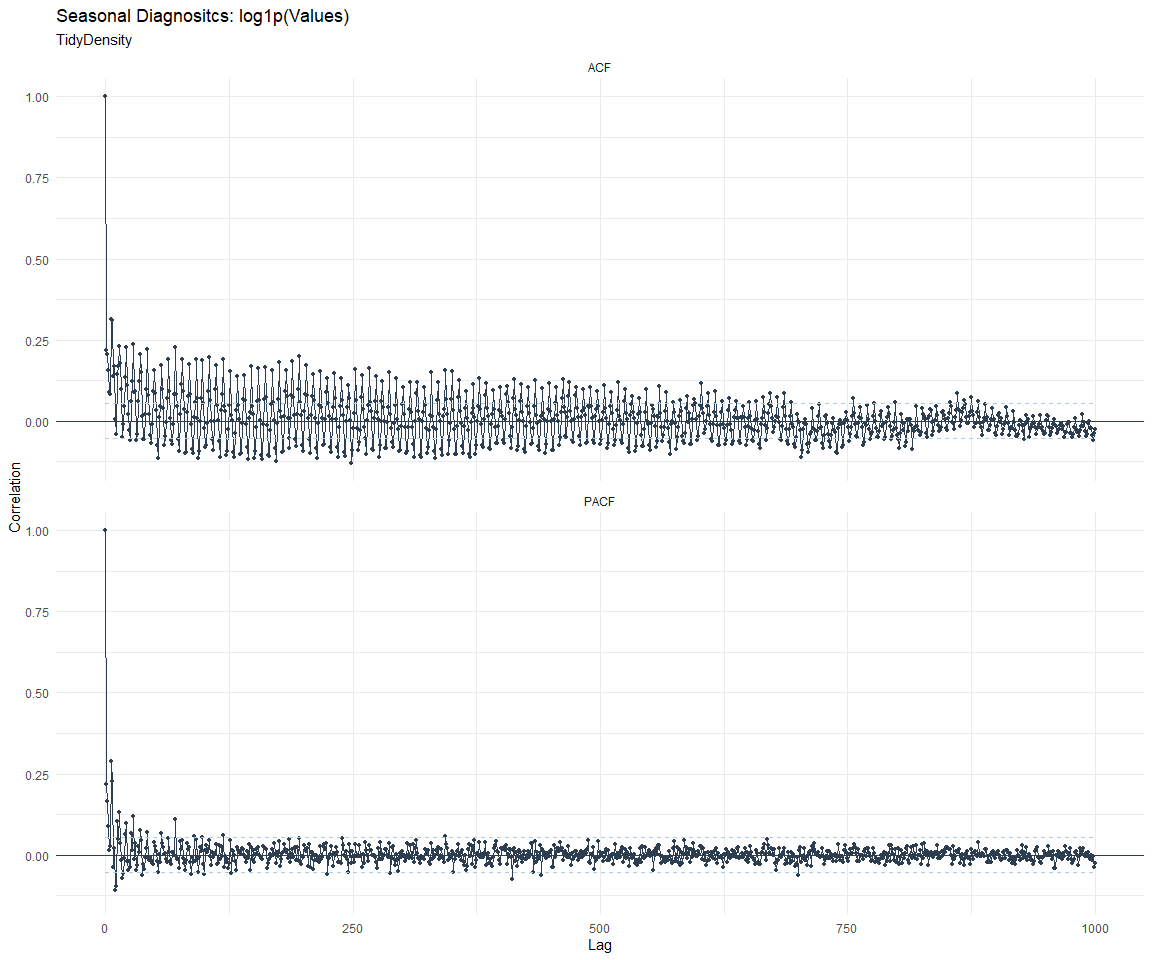
[[3]]
[[4]]
[[5]]
[[6]]
[[7]]
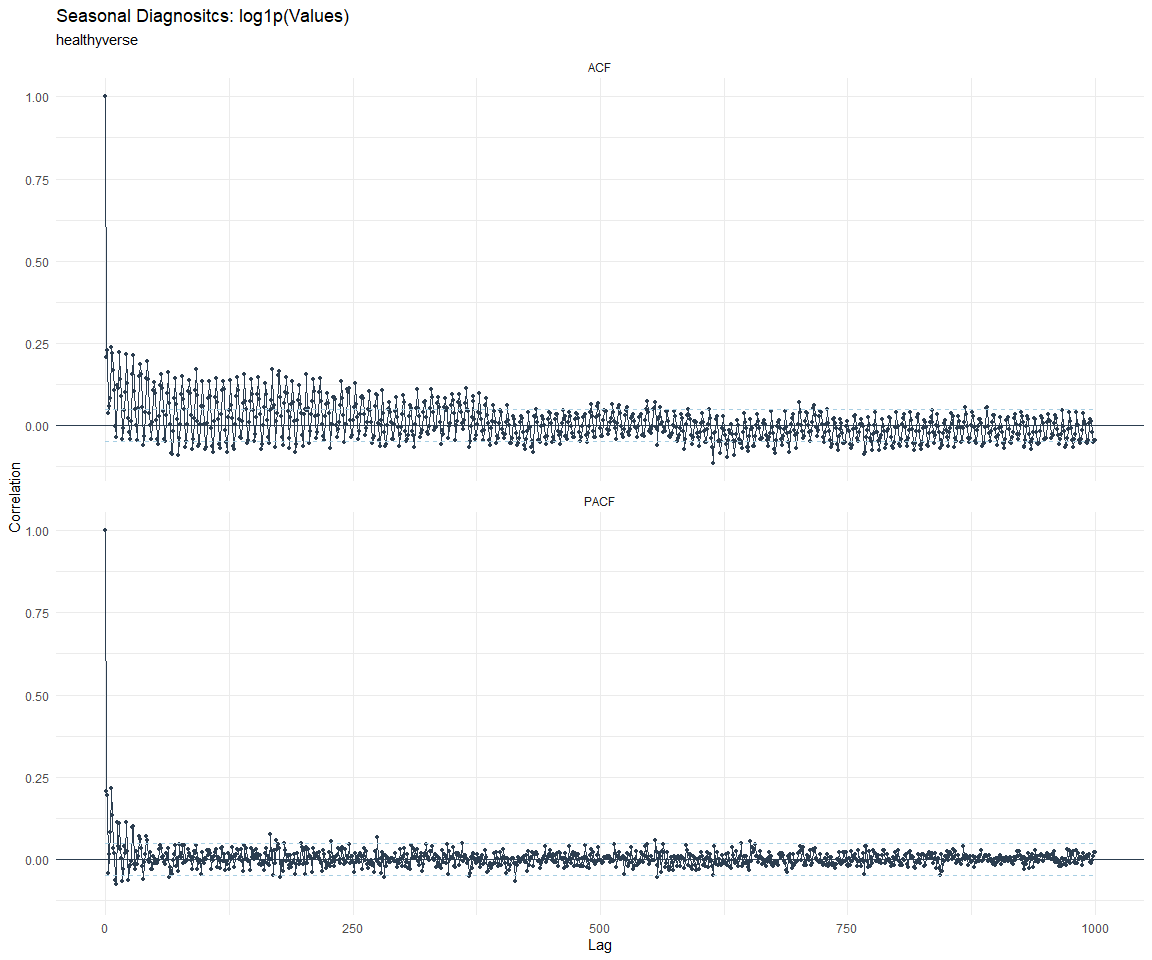
[[8]]
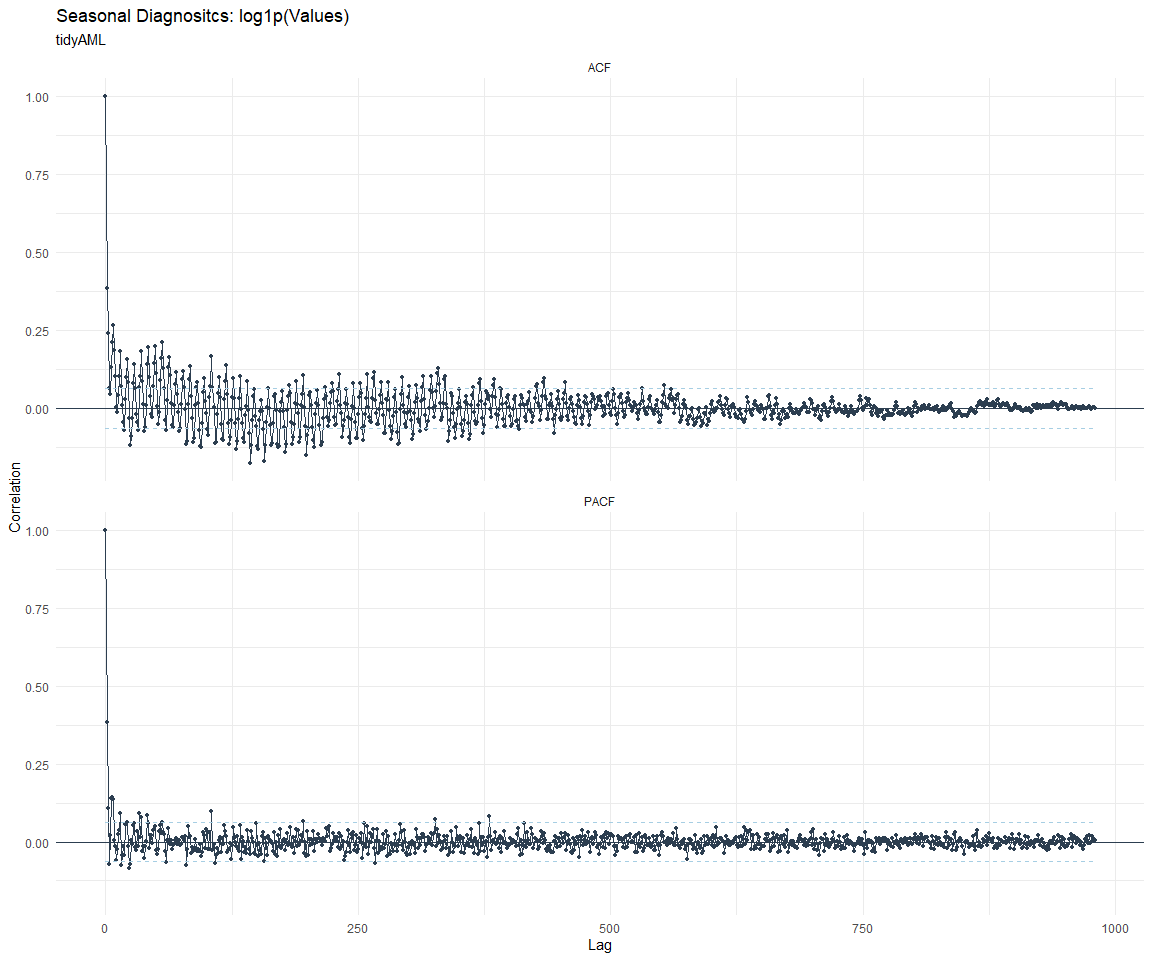
Feature Engineering
Now that we have our basic data and a shot of what it looks like, let’s
add some features to our data which can be very helpful in modeling.
Lets start by making a tibble that is aggregated by the day and
package, as we are going to be interested in forecasting the next 4
weeks or 28 days for each package. First lets get our base data.
Call:
stats::lm(formula = .formula, data = df)
Residuals:
Min 1Q Median 3Q Max
-149.85 -36.96 -11.40 27.09 824.90
Coefficients:
Estimate Std. Error
(Intercept) -1.566e+02 5.708e+01
date 9.842e-03 3.022e-03
lag(value, 1) 1.013e-01 2.286e-02
lag(value, 7) 8.732e-02 2.360e-02
lag(value, 14) 7.512e-02 2.356e-02
lag(value, 21) 8.389e-02 2.368e-02
lag(value, 28) 6.586e-02 2.359e-02
lag(value, 35) 5.114e-02 2.361e-02
lag(value, 42) 7.123e-02 2.372e-02
lag(value, 49) 6.961e-02 2.365e-02
month(date, label = TRUE).L -8.516e+00 4.819e+00
month(date, label = TRUE).Q -1.337e+00 4.745e+00
month(date, label = TRUE).C -1.454e+01 4.795e+00
month(date, label = TRUE)^4 -7.388e+00 4.843e+00
month(date, label = TRUE)^5 -5.990e+00 4.832e+00
month(date, label = TRUE)^6 7.627e-01 4.878e+00
month(date, label = TRUE)^7 -4.272e+00 4.834e+00
month(date, label = TRUE)^8 -4.209e+00 4.819e+00
month(date, label = TRUE)^9 2.881e+00 4.836e+00
month(date, label = TRUE)^10 1.040e+00 4.853e+00
month(date, label = TRUE)^11 -4.250e+00 4.840e+00
fourier_vec(date, type = "sin", K = 1, period = 7) -1.078e+01 2.169e+00
fourier_vec(date, type = "cos", K = 1, period = 7) 7.139e+00 2.244e+00
t value Pr(>|t|)
(Intercept) -2.744 0.006130 **
date 3.257 0.001146 **
lag(value, 1) 4.431 9.92e-06 ***
lag(value, 7) 3.701 0.000221 ***
lag(value, 14) 3.188 0.001456 **
lag(value, 21) 3.542 0.000407 ***
lag(value, 28) 2.791 0.005302 **
lag(value, 35) 2.166 0.030423 *
lag(value, 42) 3.003 0.002711 **
lag(value, 49) 2.944 0.003283 **
month(date, label = TRUE).L -1.767 0.077341 .
month(date, label = TRUE).Q -0.282 0.778072
month(date, label = TRUE).C -3.033 0.002453 **
month(date, label = TRUE)^4 -1.525 0.127337
month(date, label = TRUE)^5 -1.240 0.215293
month(date, label = TRUE)^6 0.156 0.875764
month(date, label = TRUE)^7 -0.884 0.376936
month(date, label = TRUE)^8 -0.874 0.382485
month(date, label = TRUE)^9 0.596 0.551361
month(date, label = TRUE)^10 0.214 0.830293
month(date, label = TRUE)^11 -0.878 0.379997
fourier_vec(date, type = "sin", K = 1, period = 7) -4.970 7.32e-07 ***
fourier_vec(date, type = "cos", K = 1, period = 7) 3.182 0.001489 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 59.38 on 1821 degrees of freedom
(49 observations deleted due to missingness)
Multiple R-squared: 0.2209, Adjusted R-squared: 0.2114
F-statistic: 23.46 on 22 and 1821 DF, p-value: < 2.2e-16
NNS Forecasting
This is something I have been wanting to try for a while. The NNS
package is a great package for forecasting time series data.
library(NNS)
data_list <- base_data |>
select(package, value) |>
group_split(package)
data_list |>
imap(
\(x, idx) {
obj <- x
x <- obj |> pull(value) |> tail(7*52)
train_set_size <- length(x) - 56
pkg <- obj |> pluck(1) |> unique()
# sf <- NNS.seas(x, modulo = 7, plot = FALSE)$periods
seas <- t(
sapply(
1:25,
function(i) c(
i,
sqrt(
mean((
NNS.ARMA(x,
h = 28,
training.set = train_set_size,
method = "lin",
seasonal.factor = i,
plot=FALSE
) - tail(x, 28)) ^ 2)))
)
)
colnames(seas) <- c("Period", "RMSE")
sf <- seas[which.min(seas[, 2]), 1]
cat(paste0("Package: ", pkg, "\n"))
NNS.ARMA.optim(
variable = x,
h = 28,
training.set = train_set_size,
#seasonal.factor = seq(12, 60, 7),
seasonal.factor = sf,
pred.int = 0.95,
plot = TRUE
)
title(
sub = paste0("\n",
"Package: ", pkg, " - NNS Optimization")
)
}
)
Package: healthyR
[1] "CURRNET METHOD: lin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'lin' , seasonal.factor = c( 3 ) ...)"
[1] "CURRENT lin OBJECTIVE FUNCTION = 15.3570404570489"
[1] "BEST method = 'lin' PATH MEMBER = c( 3 )"
[1] "BEST lin OBJECTIVE FUNCTION = 15.3570404570489"
[1] "CURRNET METHOD: nonlin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'nonlin' , seasonal.factor = c( 3 ) ...)"
[1] "CURRENT nonlin OBJECTIVE FUNCTION = 28.8412019567967"
[1] "BEST method = 'nonlin' PATH MEMBER = c( 3 )"
[1] "BEST nonlin OBJECTIVE FUNCTION = 28.8412019567967"
[1] "CURRNET METHOD: both"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'both' , seasonal.factor = c( 3 ) ...)"
[1] "CURRENT both OBJECTIVE FUNCTION = 59.1570971741622"
[1] "BEST method = 'both' PATH MEMBER = c( 3 )"
[1] "BEST both OBJECTIVE FUNCTION = 59.1570971741622"
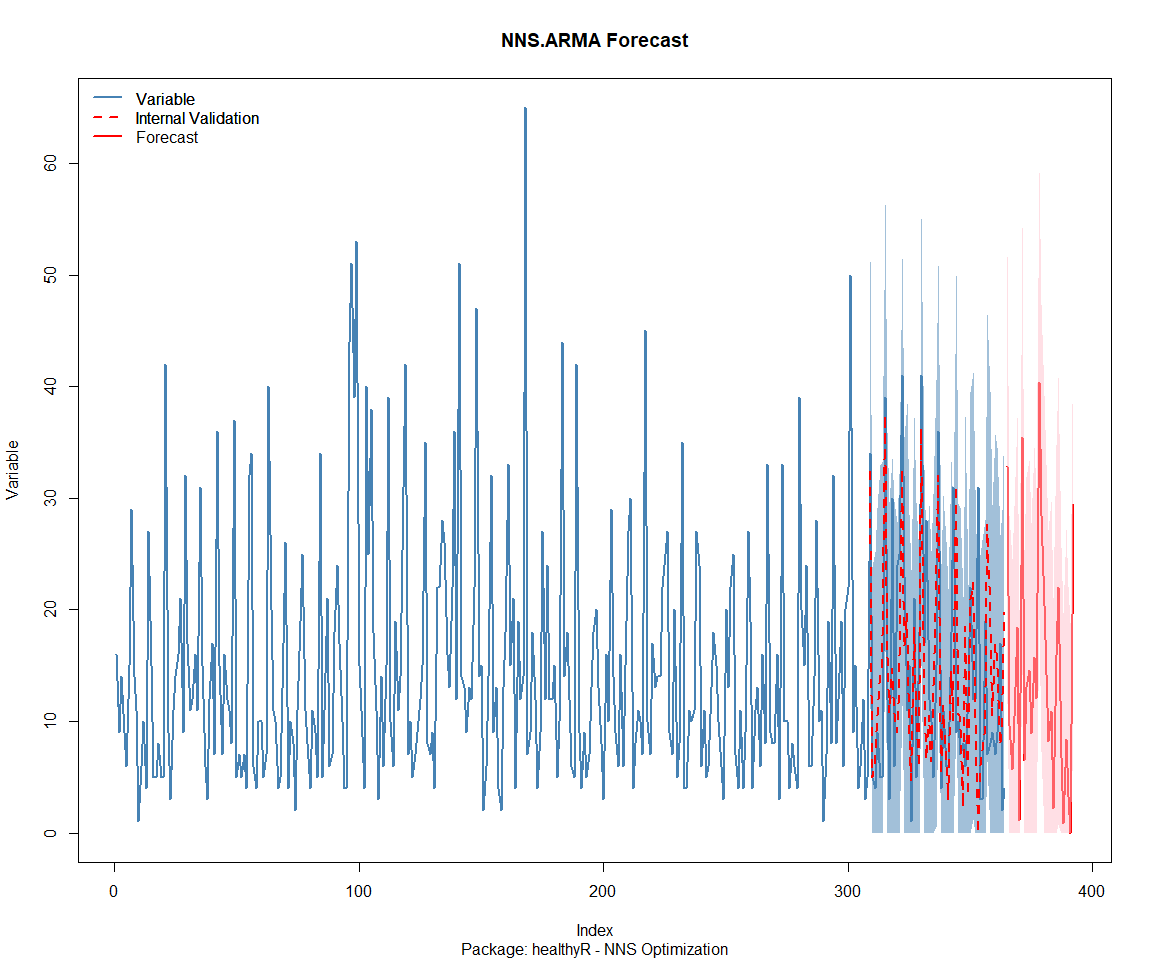
Package: healthyR.ai
[1] "CURRNET METHOD: lin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'lin' , seasonal.factor = c( 1 ) ...)"
[1] "CURRENT lin OBJECTIVE FUNCTION = 276.933666504492"
[1] "BEST method = 'lin' PATH MEMBER = c( 1 )"
[1] "BEST lin OBJECTIVE FUNCTION = 276.933666504492"
[1] "CURRNET METHOD: nonlin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'nonlin' , seasonal.factor = c( 1 ) ...)"
[1] "CURRENT nonlin OBJECTIVE FUNCTION = 35.2755205955493"
[1] "BEST method = 'nonlin' PATH MEMBER = c( 1 )"
[1] "BEST nonlin OBJECTIVE FUNCTION = 35.2755205955493"
[1] "CURRNET METHOD: both"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'both' , seasonal.factor = c( 1 ) ...)"
[1] "CURRENT both OBJECTIVE FUNCTION = 273.888850383731"
[1] "BEST method = 'both' PATH MEMBER = c( 1 )"
[1] "BEST both OBJECTIVE FUNCTION = 273.888850383731"
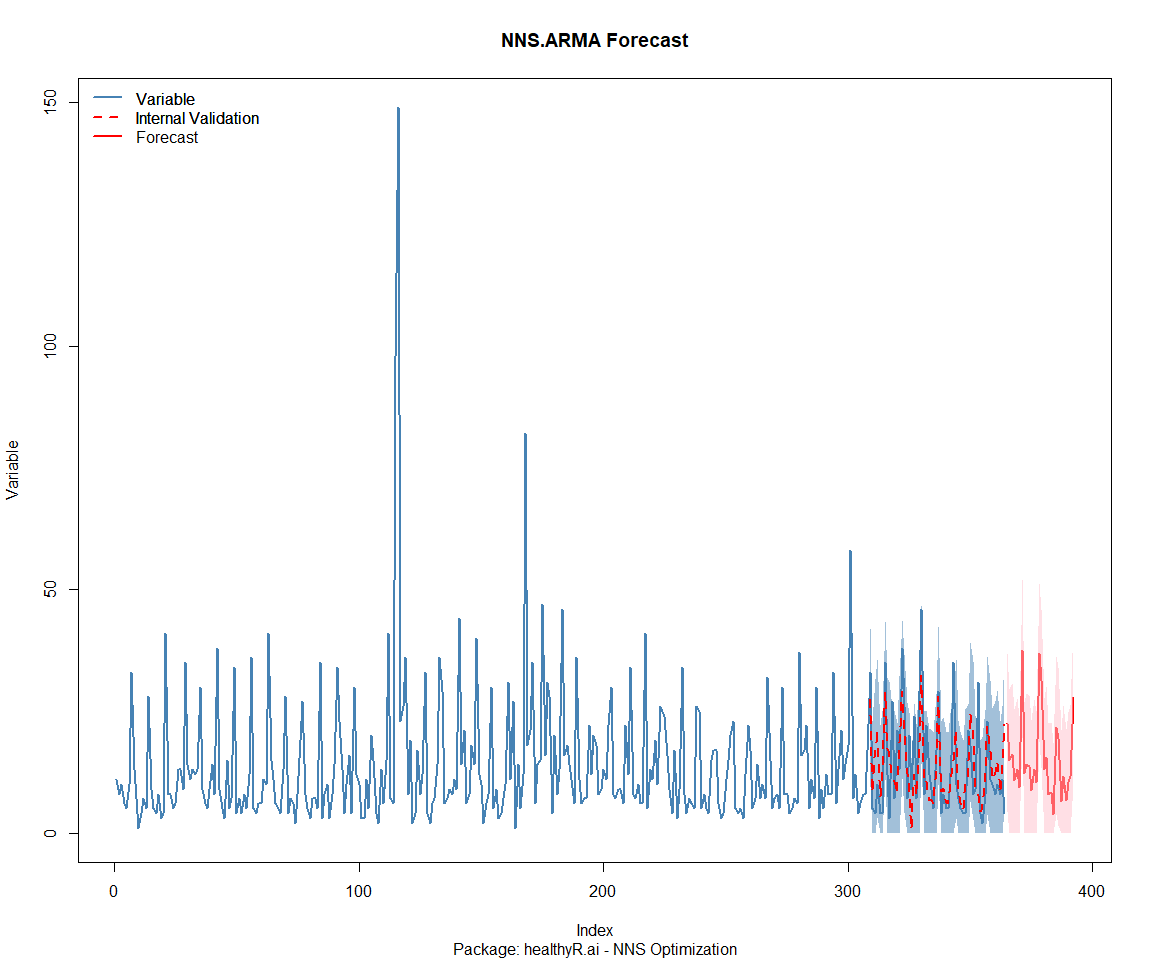
Package: healthyR.data
[1] "CURRNET METHOD: lin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'lin' , seasonal.factor = c( 14 ) ...)"
[1] "CURRENT lin OBJECTIVE FUNCTION = 7.51621261246931"
[1] "BEST method = 'lin' PATH MEMBER = c( 14 )"
[1] "BEST lin OBJECTIVE FUNCTION = 7.51621261246931"
[1] "CURRNET METHOD: nonlin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'nonlin' , seasonal.factor = c( 14 ) ...)"
[1] "CURRENT nonlin OBJECTIVE FUNCTION = 12.2893462701429"
[1] "BEST method = 'nonlin' PATH MEMBER = c( 14 )"
[1] "BEST nonlin OBJECTIVE FUNCTION = 12.2893462701429"
[1] "CURRNET METHOD: both"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'both' , seasonal.factor = c( 14 ) ...)"
[1] "CURRENT both OBJECTIVE FUNCTION = 13.9783308126132"
[1] "BEST method = 'both' PATH MEMBER = c( 14 )"
[1] "BEST both OBJECTIVE FUNCTION = 13.9783308126132"
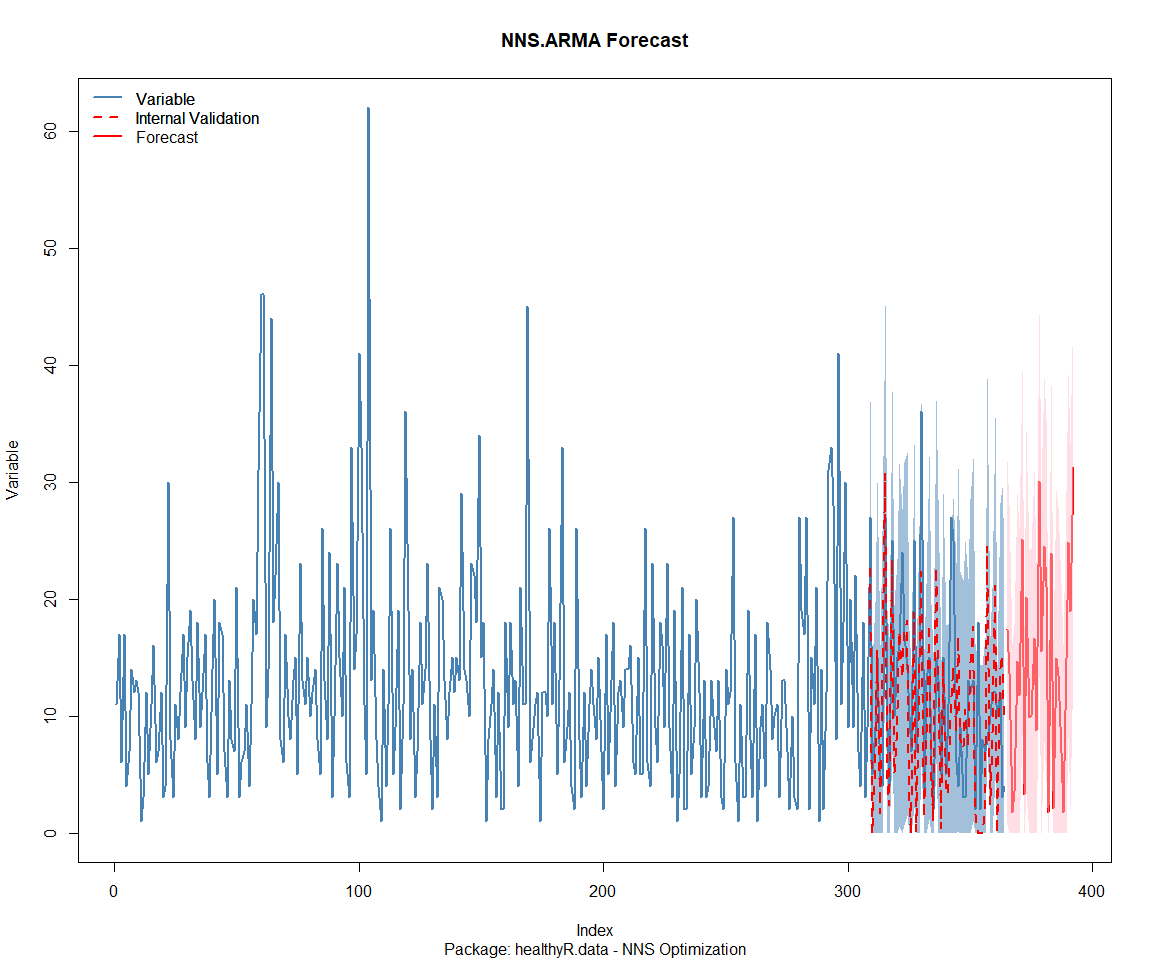
Package: healthyR.ts
[1] "CURRNET METHOD: lin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'lin' , seasonal.factor = c( 1 ) ...)"
[1] "CURRENT lin OBJECTIVE FUNCTION = 358.25180809171"
[1] "BEST method = 'lin' PATH MEMBER = c( 1 )"
[1] "BEST lin OBJECTIVE FUNCTION = 358.25180809171"
[1] "CURRNET METHOD: nonlin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'nonlin' , seasonal.factor = c( 1 ) ...)"
[1] "CURRENT nonlin OBJECTIVE FUNCTION = 108.0439549276"
[1] "BEST method = 'nonlin' PATH MEMBER = c( 1 )"
[1] "BEST nonlin OBJECTIVE FUNCTION = 108.0439549276"
[1] "CURRNET METHOD: both"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'both' , seasonal.factor = c( 1 ) ...)"
[1] "CURRENT both OBJECTIVE FUNCTION = 136.262138843649"
[1] "BEST method = 'both' PATH MEMBER = c( 1 )"
[1] "BEST both OBJECTIVE FUNCTION = 136.262138843649"
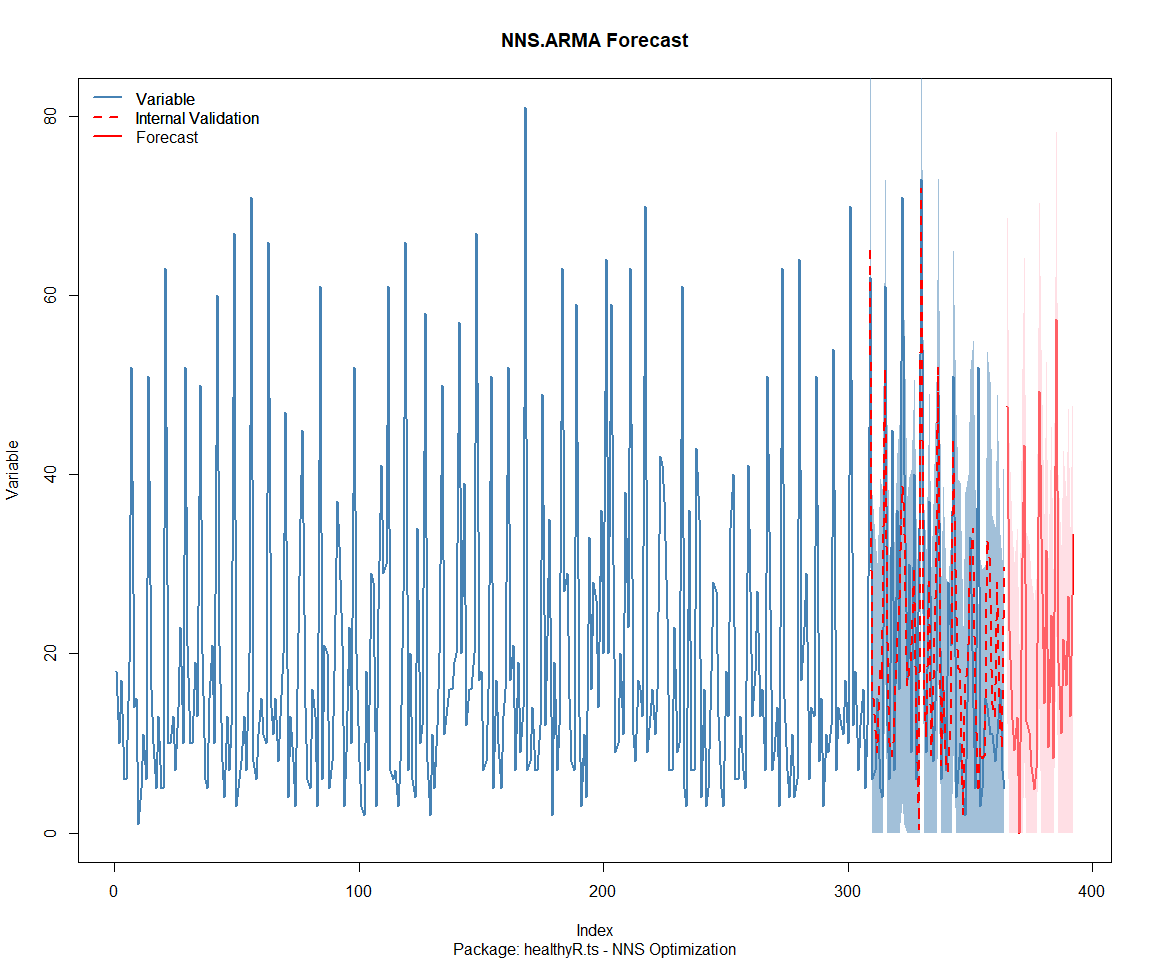
Package: healthyverse
[1] "CURRNET METHOD: lin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'lin' , seasonal.factor = c( 3 ) ...)"
[1] "CURRENT lin OBJECTIVE FUNCTION = 49.9390921183822"
[1] "BEST method = 'lin' PATH MEMBER = c( 3 )"
[1] "BEST lin OBJECTIVE FUNCTION = 49.9390921183822"
[1] "CURRNET METHOD: nonlin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'nonlin' , seasonal.factor = c( 3 ) ...)"
[1] "CURRENT nonlin OBJECTIVE FUNCTION = 5.1779777313527"
[1] "BEST method = 'nonlin' PATH MEMBER = c( 3 )"
[1] "BEST nonlin OBJECTIVE FUNCTION = 5.1779777313527"
[1] "CURRNET METHOD: both"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'both' , seasonal.factor = c( 3 ) ...)"
[1] "CURRENT both OBJECTIVE FUNCTION = 6.96652433412496"
[1] "BEST method = 'both' PATH MEMBER = c( 3 )"
[1] "BEST both OBJECTIVE FUNCTION = 6.96652433412496"
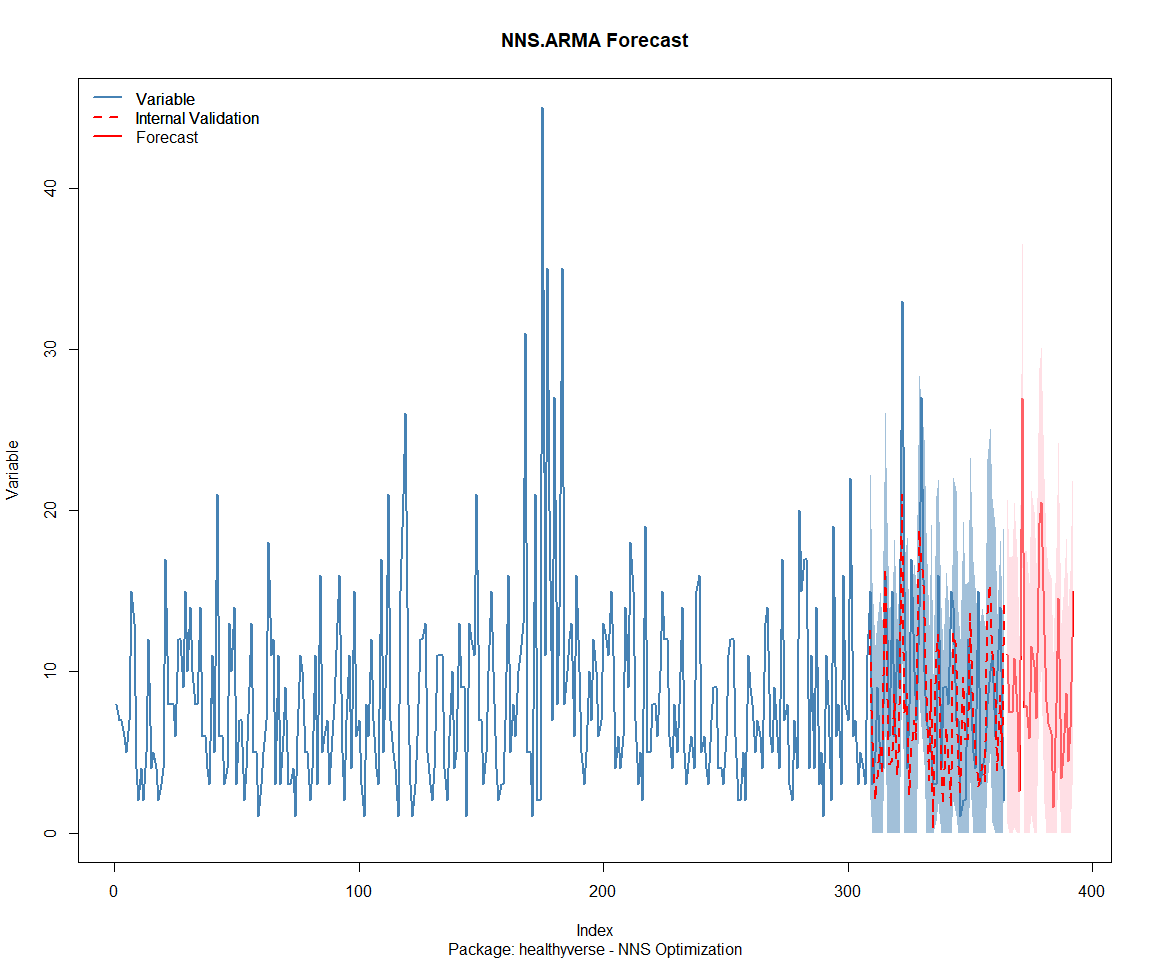
Package: RandomWalker
[1] "CURRNET METHOD: lin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'lin' , seasonal.factor = c( 16 ) ...)"
[1] "CURRENT lin OBJECTIVE FUNCTION = 22.6009268575162"
[1] "BEST method = 'lin' PATH MEMBER = c( 16 )"
[1] "BEST lin OBJECTIVE FUNCTION = 22.6009268575162"
[1] "CURRNET METHOD: nonlin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'nonlin' , seasonal.factor = c( 16 ) ...)"
[1] "CURRENT nonlin OBJECTIVE FUNCTION = 10.1745760778331"
[1] "BEST method = 'nonlin' PATH MEMBER = c( 16 )"
[1] "BEST nonlin OBJECTIVE FUNCTION = 10.1745760778331"
[1] "CURRNET METHOD: both"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'both' , seasonal.factor = c( 16 ) ...)"
[1] "CURRENT both OBJECTIVE FUNCTION = 13.6680583425527"
[1] "BEST method = 'both' PATH MEMBER = c( 16 )"
[1] "BEST both OBJECTIVE FUNCTION = 13.6680583425527"
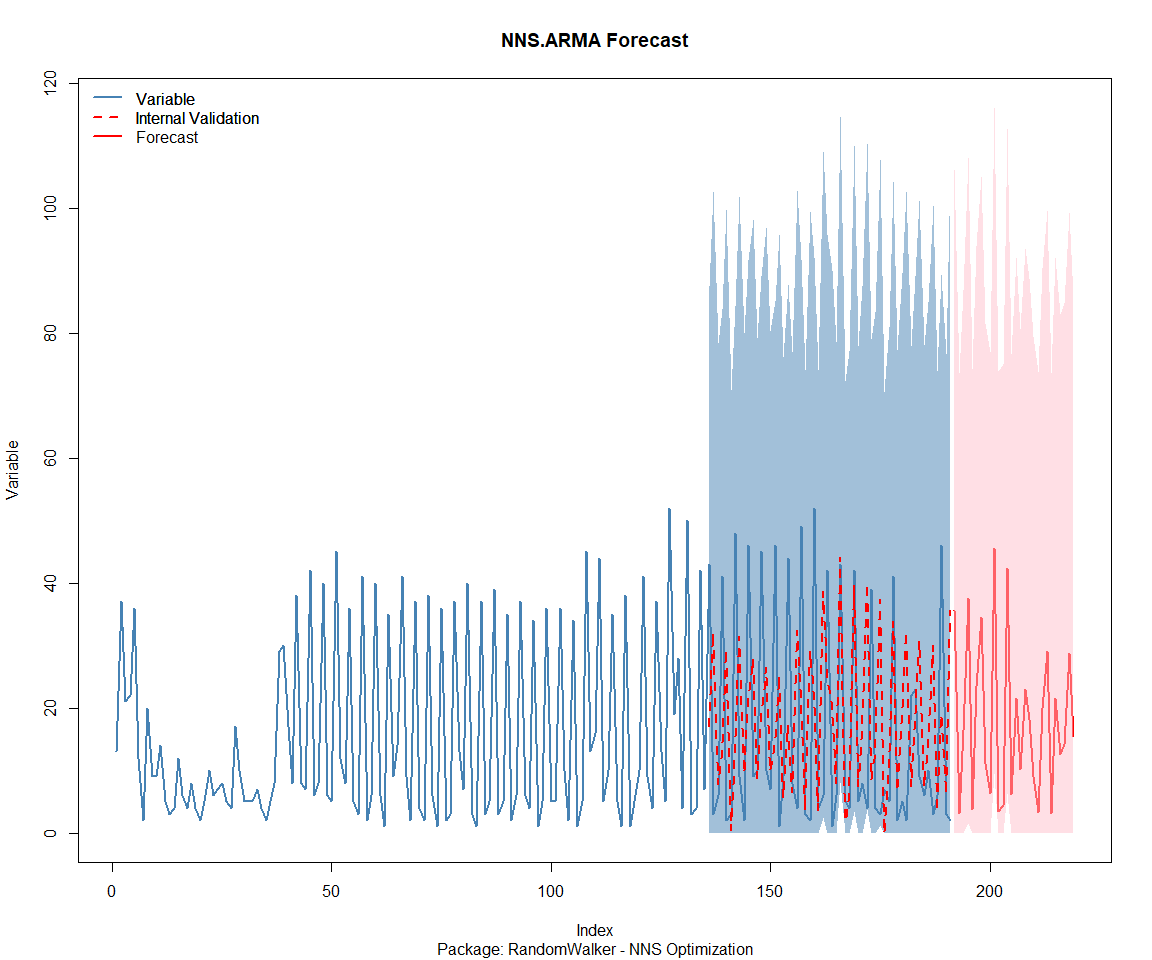
Package: tidyAML
[1] "CURRNET METHOD: lin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'lin' , seasonal.factor = c( 3 ) ...)"
[1] "CURRENT lin OBJECTIVE FUNCTION = 19.3979553516476"
[1] "BEST method = 'lin' PATH MEMBER = c( 3 )"
[1] "BEST lin OBJECTIVE FUNCTION = 19.3979553516476"
[1] "CURRNET METHOD: nonlin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'nonlin' , seasonal.factor = c( 3 ) ...)"
[1] "CURRENT nonlin OBJECTIVE FUNCTION = 5.29632296199073"
[1] "BEST method = 'nonlin' PATH MEMBER = c( 3 )"
[1] "BEST nonlin OBJECTIVE FUNCTION = 5.29632296199073"
[1] "CURRNET METHOD: both"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'both' , seasonal.factor = c( 3 ) ...)"
[1] "CURRENT both OBJECTIVE FUNCTION = 9.25351853574279"
[1] "BEST method = 'both' PATH MEMBER = c( 3 )"
[1] "BEST both OBJECTIVE FUNCTION = 9.25351853574279"
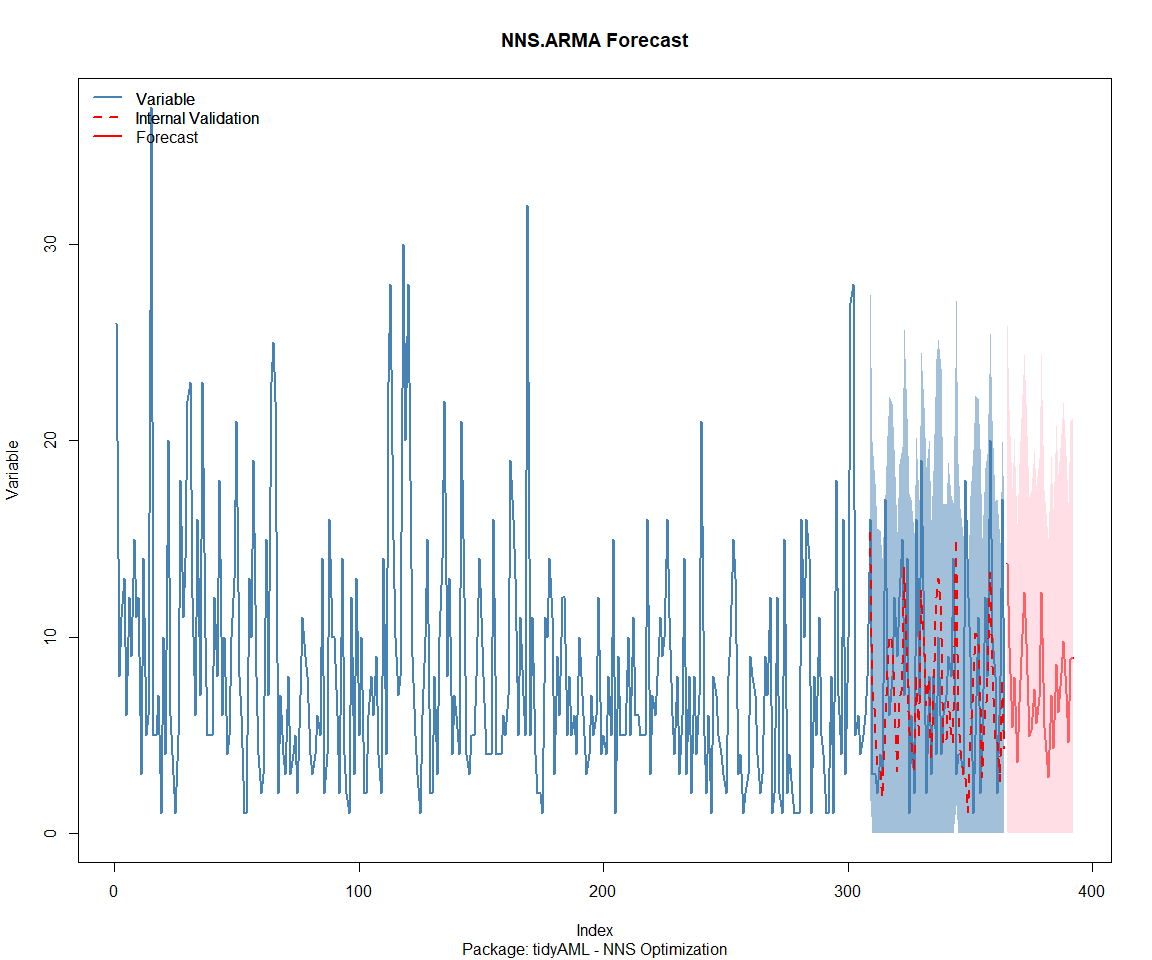
Package: TidyDensity
[1] "CURRNET METHOD: lin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'lin' , seasonal.factor = c( 3 ) ...)"
[1] "CURRENT lin OBJECTIVE FUNCTION = 7.07041635786567"
[1] "BEST method = 'lin' PATH MEMBER = c( 3 )"
[1] "BEST lin OBJECTIVE FUNCTION = 7.07041635786567"
[1] "CURRNET METHOD: nonlin"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'nonlin' , seasonal.factor = c( 3 ) ...)"
[1] "CURRENT nonlin OBJECTIVE FUNCTION = 3.60998534655865"
[1] "BEST method = 'nonlin' PATH MEMBER = c( 3 )"
[1] "BEST nonlin OBJECTIVE FUNCTION = 3.60998534655865"
[1] "CURRNET METHOD: both"
[1] "COPY LATEST PARAMETERS DIRECTLY FOR NNS.ARMA() IF ERROR:"
[1] "NNS.ARMA(... method = 'both' , seasonal.factor = c( 3 ) ...)"
[1] "CURRENT both OBJECTIVE FUNCTION = 4.18407714635079"
[1] "BEST method = 'both' PATH MEMBER = c( 3 )"
[1] "BEST both OBJECTIVE FUNCTION = 4.18407714635079"
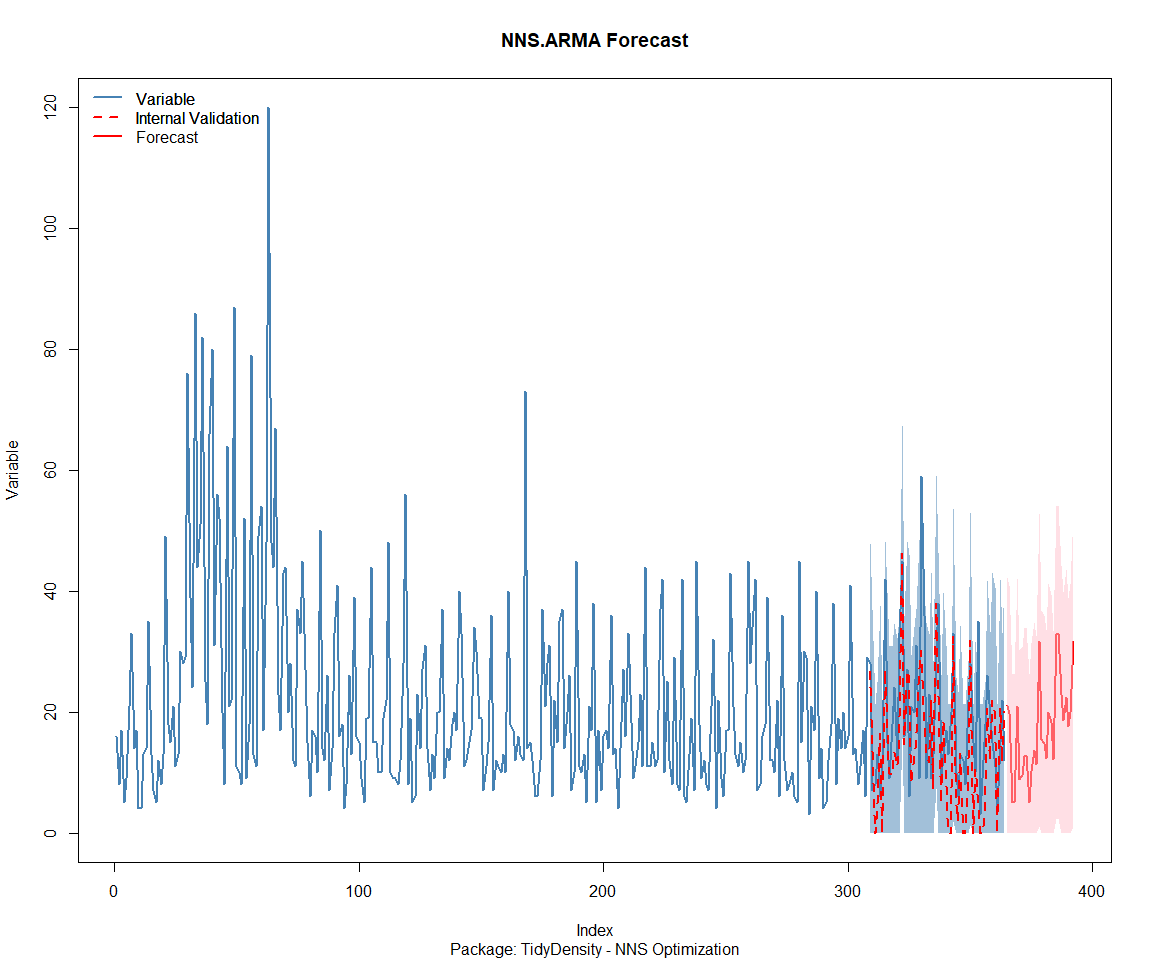
[[1]]
NULL
[[2]]
NULL
[[3]]
NULL
[[4]]
NULL
[[5]]
NULL
[[6]]
NULL
[[7]]
NULL
[[8]]
NULL
Pre-Processing
Now we are going to do some basic pre-processing.
data_padded_tbl <- base_data %>%
pad_by_time(
.date_var = date,
.pad_value = 0
)
# Get log interval and standardization parameters
log_params <- liv(data_padded_tbl$value, limit_lower = 0, offset = 1, silent = TRUE)
limit_lower <- log_params$limit_lower
limit_upper <- log_params$limit_upper
offset <- log_params$offset
data_liv_tbl <- data_padded_tbl %>%
# Get log interval transform
mutate(value_trans = liv(value, limit_lower = 0, offset = 1, silent = TRUE)$log_scaled)
# Get Standardization Params
std_params <- standard_vec(data_liv_tbl$value_trans, silent = TRUE)
std_mean <- std_params$mean
std_sd <- std_params$sd
data_transformed_tbl <- data_liv_tbl %>%
group_by(package) %>%
# get standardization
mutate(value_trans = standard_vec(value_trans, silent = TRUE)$standard_scaled) %>%
tk_augment_fourier(
.date_var = date,
.periods = c(7, 14, 30, 90, 180),
.K = 2
) %>%
tk_augment_timeseries_signature(
.date_var = date
) %>%
ungroup() %>%
select(-c(value, -year.iso))
Since this is panel data we can follow one of two different modeling strategies. We can search for a global model in the panel data or we can use nested forecasting finding the best model for each of the time series. Since we only have 5 panels, we will use nested forecasting.
To do this we will use the nest_timeseries and
split_nested_timeseries functions to create a nested tibble.
horizon <- 4*7
nested_data_tbl <- data_transformed_tbl %>%
# 0. Filter out column where package is NA
filter(!is.na(package)) %>%
# 1. Extending: We'll predict n days into the future.
extend_timeseries(
.id_var = package,
.date_var = date,
.length_future = horizon
) %>%
# 2. Nesting: We'll group by id, and create a future dataset
# that forecasts n days of extended data and
# an actual dataset that contains n*2 days
nest_timeseries(
.id_var = package,
.length_future = horizon
#.length_actual = horizon*2
) %>%
# 3. Splitting: We'll take the actual data and create splits
# for accuracy and confidence interval estimation of n das (test)
# and the rest is training data
split_nested_timeseries(
.length_test = horizon
)
nested_data_tbl
# A tibble: 8 × 4
package .actual_data .future_data .splits
<fct> <list> <list> <list>
1 healthyR.data <tibble [1,883 × 50]> <tibble [28 × 50]> <split [1855|28]>
2 healthyR <tibble [1,876 × 50]> <tibble [28 × 50]> <split [1848|28]>
3 healthyR.ts <tibble [1,812 × 50]> <tibble [28 × 50]> <split [1784|28]>
4 healthyverse <tibble [1,774 × 50]> <tibble [28 × 50]> <split [1746|28]>
5 healthyR.ai <tibble [1,618 × 50]> <tibble [28 × 50]> <split [1590|28]>
6 TidyDensity <tibble [1,469 × 50]> <tibble [28 × 50]> <split [1441|28]>
7 tidyAML <tibble [1,076 × 50]> <tibble [28 × 50]> <split [1048|28]>
8 RandomWalker <tibble [499 × 50]> <tibble [28 × 50]> <split [471|28]>
Now it is time to make some recipes and models using the modeltime workflow.
Modeltime Workflow
Recipe Object
recipe_base <- recipe(
value_trans ~ .
, data = extract_nested_test_split(nested_data_tbl)
)
recipe_base
recipe_date <- recipe(
value_trans ~ date
, data = extract_nested_test_split(nested_data_tbl)
)
Models
# Models ------------------------------------------------------------------
# Auto ARIMA --------------------------------------------------------------
model_spec_arima_no_boost <- arima_reg() %>%
set_engine(engine = "auto_arima")
wflw_auto_arima <- workflow() %>%
add_recipe(recipe = recipe_date) %>%
add_model(model_spec_arima_no_boost)
# NNETAR ------------------------------------------------------------------
model_spec_nnetar <- nnetar_reg(
mode = "regression"
, seasonal_period = "auto"
) %>%
set_engine("nnetar")
wflw_nnetar <- workflow() %>%
add_recipe(recipe = recipe_base) %>%
add_model(model_spec_nnetar)
# TSLM --------------------------------------------------------------------
model_spec_lm <- linear_reg() %>%
set_engine("lm")
wflw_lm <- workflow() %>%
add_recipe(recipe = recipe_base) %>%
add_model(model_spec_lm)
# MARS --------------------------------------------------------------------
model_spec_mars <- mars(mode = "regression") %>%
set_engine("earth")
wflw_mars <- workflow() %>%
add_recipe(recipe = recipe_date) %>%
add_model(model_spec_mars)
Nested Modeltime Tables
nested_modeltime_tbl <- modeltime_nested_fit(
# Nested Data
nested_data = nested_data_tbl,
control = control_nested_fit(
verbose = TRUE,
allow_par = FALSE
),
# Add workflows
wflw_auto_arima,
wflw_lm,
wflw_mars,
wflw_nnetar
)
nested_modeltime_tbl <- nested_modeltime_tbl[!is.na(nested_modeltime_tbl$package),]
Model Accuracy
nested_modeltime_tbl %>%
extract_nested_test_accuracy() %>%
filter(!is.na(package)) %>%
knitr::kable()
| package | .model_id | .model_desc | .type | mae | mape | mase | smape | rmse | rsq |
|---|---|---|---|---|---|---|---|---|---|
| healthyR.data | 1 | ARIMA | Test | 0.6099053 | 117.21420 | 0.7722328 | 128.90776 | 0.8372174 | 0.0105638 |
| healthyR.data | 2 | LM | Test | 0.5905745 | 123.58708 | 0.7477571 | 147.79374 | 0.7827217 | 0.0340390 |
| healthyR.data | 3 | EARTH | Test | 0.6019733 | 135.07539 | 0.7621898 | 127.08442 | 0.8294939 | 0.0209266 |
| healthyR.data | 4 | NNAR | Test | 0.6722387 | 158.41915 | 0.8511564 | 154.81544 | 0.8590343 | 0.0008008 |
| healthyR | 1 | ARIMA | Test | 0.6737251 | 546.80869 | 0.5959695 | 126.88334 | 0.9054046 | 0.0334309 |
| healthyR | 2 | LM | Test | 0.6699080 | 893.82905 | 0.5925929 | 126.07154 | 0.8948428 | 0.0604676 |
| healthyR | 3 | EARTH | Test | 0.6279548 | 862.74587 | 0.5554816 | 107.23046 | 0.8611065 | 0.0027090 |
| healthyR | 4 | NNAR | Test | 0.7138218 | 836.05293 | 0.6314386 | 139.40973 | 0.9167976 | 0.0451527 |
| healthyR.ts | 1 | ARIMA | Test | 1.0257314 | 126.78107 | 0.7765851 | 163.98223 | 1.2879524 | 0.0002964 |
| healthyR.ts | 2 | LM | Test | 1.1579660 | 158.18061 | 0.8767003 | 163.82692 | 1.4202924 | 0.0251814 |
| healthyR.ts | 3 | EARTH | Test | 1.0831554 | 366.59420 | 0.8200609 | 116.44168 | 1.3294268 | 0.2744346 |
| healthyR.ts | 4 | NNAR | Test | 1.1993967 | 231.12678 | 0.9080677 | 147.72512 | 1.5289717 | 0.0871192 |
| healthyverse | 1 | ARIMA | Test | 1.2016115 | 81.93144 | 1.6803538 | 125.38022 | 1.3326350 | 0.1057826 |
| healthyverse | 2 | LM | Test | 1.1611780 | 81.10437 | 1.6238110 | 121.28604 | 1.3147848 | 0.0697262 |
| healthyverse | 3 | EARTH | Test | 3.3960190 | 337.82594 | 4.7490503 | 109.20137 | 3.6498963 | 0.1503331 |
| healthyverse | 4 | NNAR | Test | 1.0904910 | 73.50847 | 1.5249611 | 115.39467 | 1.2640252 | 0.0812698 |
| healthyR.ai | 1 | ARIMA | Test | 0.6001248 | 70.60701 | 0.9082831 | 122.24916 | 0.7548092 | 0.0016209 |
| healthyR.ai | 2 | LM | Test | 0.6700701 | 171.00100 | 1.0141446 | 137.90971 | 0.7809900 | 0.1699805 |
| healthyR.ai | 3 | EARTH | Test | 1.2926248 | 549.04055 | 1.9563751 | 102.77341 | 1.4866128 | 0.0184838 |
| healthyR.ai | 4 | NNAR | Test | 0.6987475 | 167.46169 | 1.0575476 | 143.13555 | 0.8043861 | 0.1868994 |
| TidyDensity | 1 | ARIMA | Test | 0.9565522 | 133.54624 | 0.5960831 | 182.51569 | 1.1466463 | 0.0065542 |
| TidyDensity | 2 | LM | Test | 1.0412624 | 249.56895 | 0.6488709 | 162.72273 | 1.1513916 | 0.0372852 |
| TidyDensity | 3 | EARTH | Test | 0.9034856 | 113.02951 | 0.5630143 | 130.61883 | 1.2129834 | 0.0032776 |
| TidyDensity | 4 | NNAR | Test | 0.9585424 | 146.44201 | 0.5973234 | 150.46667 | 1.1579822 | 0.0141777 |
| tidyAML | 1 | ARIMA | Test | 0.5206079 | 202.41856 | 0.6278392 | 89.96663 | 0.6981000 | 0.0723444 |
| tidyAML | 2 | LM | Test | 0.7470455 | 252.67951 | 0.9009169 | 151.71324 | 0.9319798 | 0.0470904 |
| tidyAML | 3 | EARTH | Test | 0.6843030 | 351.48822 | 0.8252511 | 99.94376 | 0.8432666 | 0.0285179 |
| tidyAML | 4 | NNAR | Test | 0.5576257 | 207.34374 | 0.6724816 | 112.59219 | 0.7818872 | 0.0224427 |
| RandomWalker | 1 | ARIMA | Test | 0.7943986 | 103.36364 | 0.5185202 | 147.02584 | 0.9248691 | 0.2565256 |
| RandomWalker | 2 | LM | Test | 0.8873900 | 108.13327 | 0.5792176 | 166.28448 | 1.0423418 | 0.0031471 |
| RandomWalker | 3 | EARTH | Test | 0.8676977 | 105.27563 | 0.5663640 | 150.49668 | 1.0482403 | 0.0043491 |
| RandomWalker | 4 | NNAR | Test | 1.0761369 | 233.46729 | 0.7024166 | 164.31963 | 1.1305924 | 0.0052111 |
Plot Models
nested_modeltime_tbl %>%
extract_nested_test_forecast() %>%
group_by(package) %>%
filter_by_time(.date_var = .index, .start_date = max(.index) - 60) %>%
ungroup() %>%
plot_modeltime_forecast(
.interactive = FALSE,
.conf_interval_show = FALSE,
.facet_scales = "free"
) +
theme_minimal() +
facet_wrap(~ package, nrow = 3) +
theme(legend.position = "bottom")

Best Model
best_nested_modeltime_tbl <- nested_modeltime_tbl %>%
modeltime_nested_select_best(
metric = "rmse",
minimize = TRUE,
filter_test_forecasts = TRUE
)
best_nested_modeltime_tbl %>%
extract_nested_best_model_report()
# Nested Modeltime Table
# A tibble: 8 × 10
package .model_id .model_desc .type mae mape mase smape rmse rsq
<fct> <int> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 healthyR.da… 2 LM Test 0.591 124. 0.748 148. 0.783 3.40e-2
2 healthyR 3 EARTH Test 0.628 863. 0.555 107. 0.861 2.71e-3
3 healthyR.ts 1 ARIMA Test 1.03 127. 0.777 164. 1.29 2.96e-4
4 healthyverse 4 NNAR Test 1.09 73.5 1.52 115. 1.26 8.13e-2
5 healthyR.ai 1 ARIMA Test 0.600 70.6 0.908 122. 0.755 1.62e-3
6 TidyDensity 1 ARIMA Test 0.957 134. 0.596 183. 1.15 6.55e-3
7 tidyAML 1 ARIMA Test 0.521 202. 0.628 90.0 0.698 7.23e-2
8 RandomWalker 1 ARIMA Test 0.794 103. 0.519 147. 0.925 2.57e-1
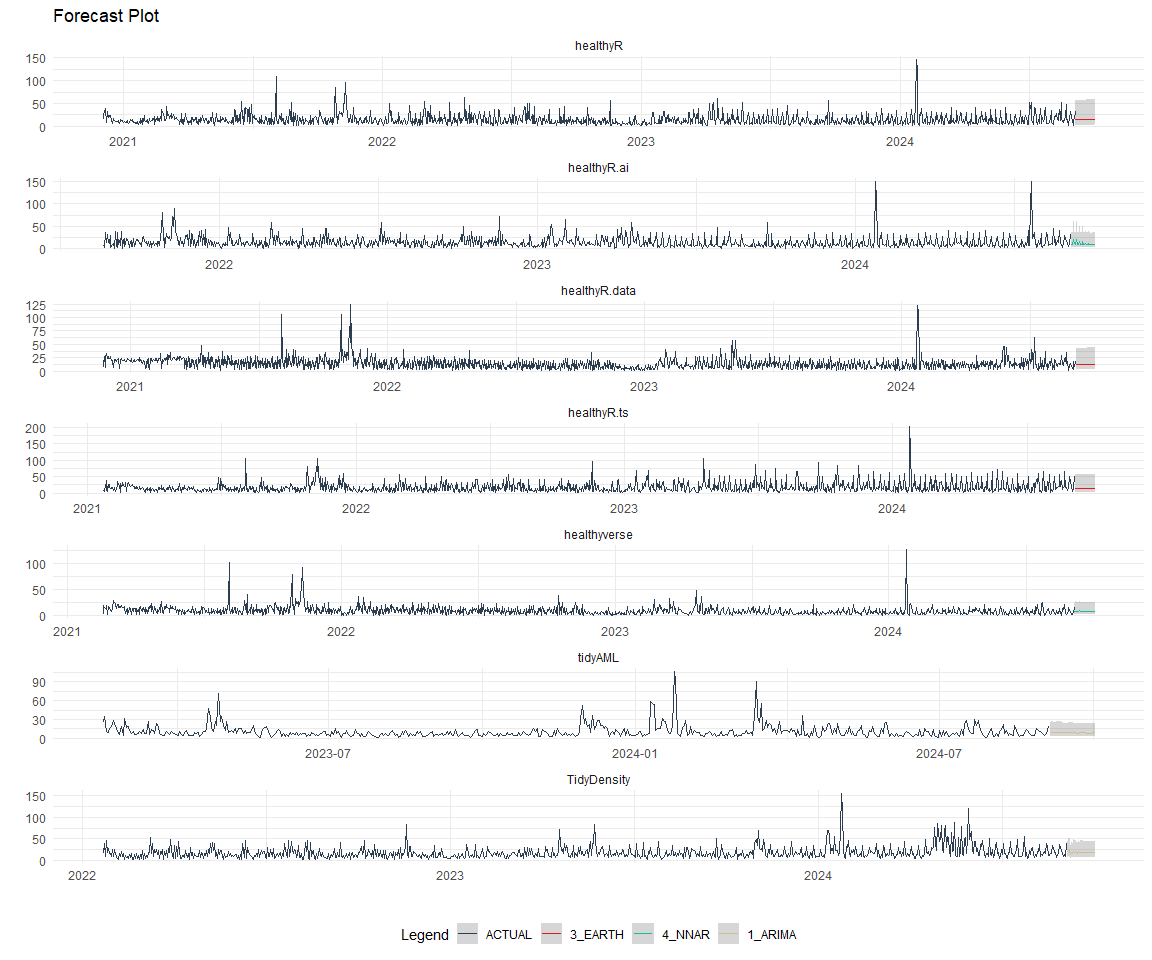
best_nested_modeltime_tbl %>%
extract_nested_test_forecast() %>%
#filter(!is.na(.model_id)) %>%
group_by(package) %>%
filter_by_time(.date_var = .index, .start_date = max(.index) - 60) %>%
ungroup() %>%
plot_modeltime_forecast(
.interactive = FALSE,
.conf_interval_alpha = 0.2,
.facet_scales = "free"
) +
facet_wrap(~ package, nrow = 3) +
theme_minimal() +
theme(legend.position = "bottom")
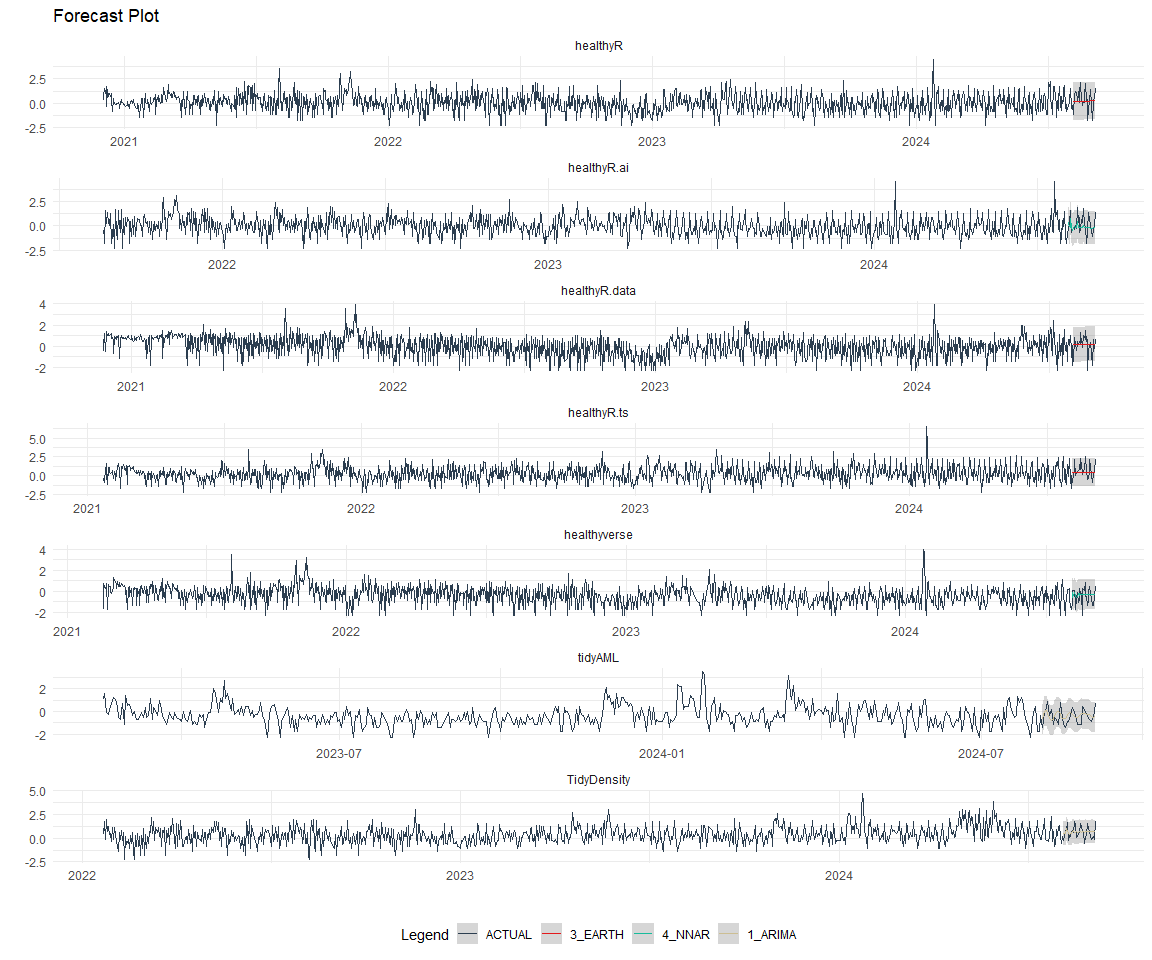
Refitting and Future Forecast
Now that we have the best models, we can make our future forecasts.
nested_modeltime_refit_tbl <- best_nested_modeltime_tbl %>%
modeltime_nested_refit(
control = control_nested_refit(verbose = TRUE)
)
nested_modeltime_refit_tbl
# Nested Modeltime Table
# A tibble: 8 × 5
package .actual_data .future_data .splits .modeltime_tables
<fct> <list> <list> <list> <list>
1 healthyR.data <tibble> <tibble> <split [1855|28]> <mdl_tm_t [1 × 5]>
2 healthyR <tibble> <tibble> <split [1848|28]> <mdl_tm_t [1 × 5]>
3 healthyR.ts <tibble> <tibble> <split [1784|28]> <mdl_tm_t [1 × 5]>
4 healthyverse <tibble> <tibble> <split [1746|28]> <mdl_tm_t [1 × 5]>
5 healthyR.ai <tibble> <tibble> <split [1590|28]> <mdl_tm_t [1 × 5]>
6 TidyDensity <tibble> <tibble> <split [1441|28]> <mdl_tm_t [1 × 5]>
7 tidyAML <tibble> <tibble> <split [1048|28]> <mdl_tm_t [1 × 5]>
8 RandomWalker <tibble> <tibble> <split [471|28]> <mdl_tm_t [1 × 5]>
nested_modeltime_refit_tbl %>%
extract_nested_future_forecast() %>%
group_by(package) %>%
mutate(across(.value:.conf_hi, .fns = ~ standard_inv_vec(
x = .,
mean = std_mean,
sd = std_sd
)$standard_inverse_value)) %>%
mutate(across(.value:.conf_hi, .fns = ~ liiv(
x = .,
limit_lower = limit_lower,
limit_upper = limit_upper,
offset = offset
)$rescaled_v)) %>%
filter_by_time(.date_var = .index, .start_date = max(.index) - 60) %>%
ungroup() %>%
plot_modeltime_forecast(
.interactive = FALSE,
.conf_interval_alpha = 0.2,
.facet_scales = "free"
) +
facet_wrap(~ package, nrow = 3) +
theme_minimal() +
theme(legend.position = "bottom")