
Estimate Negative Binomial Parameters
Source:R/est-param-negative-binomial.R
util_negative_binomial_param_estimate.RdThe function will return a list output by default, and if the parameter
.auto_gen_empirical is set to TRUE then the empirical data given to the
parameter .x will be run through the tidy_empirical() function and combined
with the estimated negative binomial data.
Three different methods of shape parameters are supplied:
MLE/MME
MMUE
MLE via
optimfunction.
Arguments
- .x
The vector of data to be passed to the function.
- .size
The size parameter, the default is 1.
- .auto_gen_empirical
This is a boolean value of TRUE/FALSE with default set to TRUE. This will automatically create the
tidy_empirical()output for the.xparameter and use thetidy_combine_distributions(). The user can then plot out the data using$combined_data_tblfrom the function output.
Details
This function will attempt to estimate the negative binomial size and prob parameters given some vector of values.
See also
Other Parameter Estimation:
util_bernoulli_param_estimate(),
util_beta_param_estimate(),
util_binomial_param_estimate(),
util_burr_param_estimate(),
util_cauchy_param_estimate(),
util_chisquare_param_estimate(),
util_exponential_param_estimate(),
util_f_param_estimate(),
util_gamma_param_estimate(),
util_generalized_beta_param_estimate(),
util_generalized_pareto_param_estimate(),
util_geometric_param_estimate(),
util_hypergeometric_param_estimate(),
util_inverse_burr_param_estimate(),
util_inverse_pareto_param_estimate(),
util_inverse_weibull_param_estimate(),
util_logistic_param_estimate(),
util_lognormal_param_estimate(),
util_normal_param_estimate(),
util_paralogistic_param_estimate(),
util_pareto1_param_estimate(),
util_pareto_param_estimate(),
util_poisson_param_estimate(),
util_t_param_estimate(),
util_triangular_param_estimate(),
util_uniform_param_estimate(),
util_weibull_param_estimate(),
util_zero_truncated_binomial_param_estimate(),
util_zero_truncated_geometric_param_estimate(),
util_zero_truncated_negative_binomial_param_estimate(),
util_zero_truncated_poisson_param_estimate()
Other Binomial:
tidy_binomial(),
tidy_negative_binomial(),
tidy_zero_truncated_binomial(),
tidy_zero_truncated_negative_binomial(),
util_binomial_param_estimate(),
util_binomial_stats_tbl(),
util_zero_truncated_binomial_param_estimate(),
util_zero_truncated_binomial_stats_tbl(),
util_zero_truncated_negative_binomial_param_estimate(),
util_zero_truncated_negative_binomial_stats_tbl()
Examples
library(dplyr)
library(ggplot2)
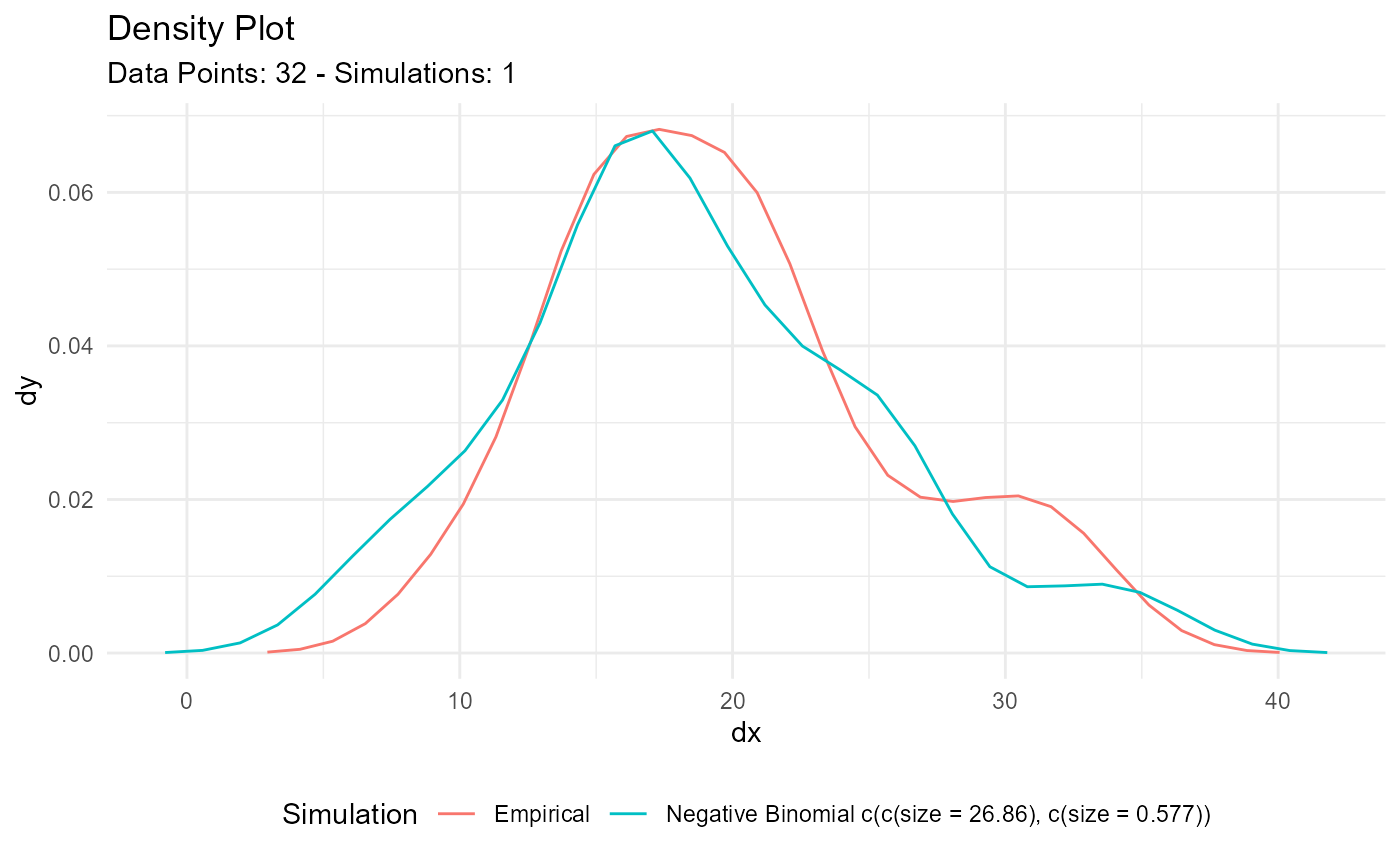
x <- as.integer(mtcars$mpg)
output <- util_negative_binomial_param_estimate(x, .size = 1)
output$parameter_tbl
#> # A tibble: 3 × 9
#> dist_type samp_size min max mean method size prob shape_ratio
#> <chr> <int> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 Negative Binomial 32 10 33 19.7 EnvSta… 32 0.0483 662
#> 2 Negative Binomial 32 10 33 19.7 EnvSta… 32 0.0469 682.
#> 3 Negative Binomial 32 10 33 19.7 MLE_Op… 26.9 0.577 46.5
output$combined_data_tbl |>
tidy_combined_autoplot()
 t <- rnbinom(50, 1, .1)
util_negative_binomial_param_estimate(t, .size = 1)$parameter_tbl
#> # A tibble: 3 × 9
#> dist_type samp_size min max mean method size prob shape_ratio
#> <chr> <int> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 Negative Binomial 50 0 48 9.42 EnvSt… 50 0.0960 521
#> 2 Negative Binomial 50 0 48 9.42 EnvSt… 50 0.0942 531.
#> 3 Negative Binomial 50 0 48 9.42 MLE_O… 0.980 0.0943 10.4
t <- rnbinom(50, 1, .1)
util_negative_binomial_param_estimate(t, .size = 1)$parameter_tbl
#> # A tibble: 3 × 9
#> dist_type samp_size min max mean method size prob shape_ratio
#> <chr> <int> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 Negative Binomial 50 0 48 9.42 EnvSt… 50 0.0960 521
#> 2 Negative Binomial 50 0 48 9.42 EnvSt… 50 0.0942 531.
#> 3 Negative Binomial 50 0 48 9.42 MLE_O… 0.980 0.0943 10.4